Overview:
CMsiRNAdb is a manually curated database containing 43,153 siRNA patent entries, including 15,760 unique chemically modified sequences derived from 90 patents, 39 cell types, 36 specific modification types, and 13 target genes. It provides comprehensive data on both sense and antisense strand sequences (original and modified versions), experimental parameters (cell models, concentration, treatment duration), efficacy metrics (inhibition rate with standard deviation), and nucleotide-level modification annotations. The database integrates advanced visualization and analytical tools to explore correlations between chemical modification patterns and gene silencing activity. Designed as a one-stop solution for researchers, CMsiRNAdb enables efficient siRNA optimization through its structured data framework and intelligent analysis capabilities.
The homepage is displayed in the following Fig 1-1:
1. Main functions of the database are provided in menu bar form (boxed in red).
2. Introduction and overview for CMsiRNAdb.
3. Quick search for users.
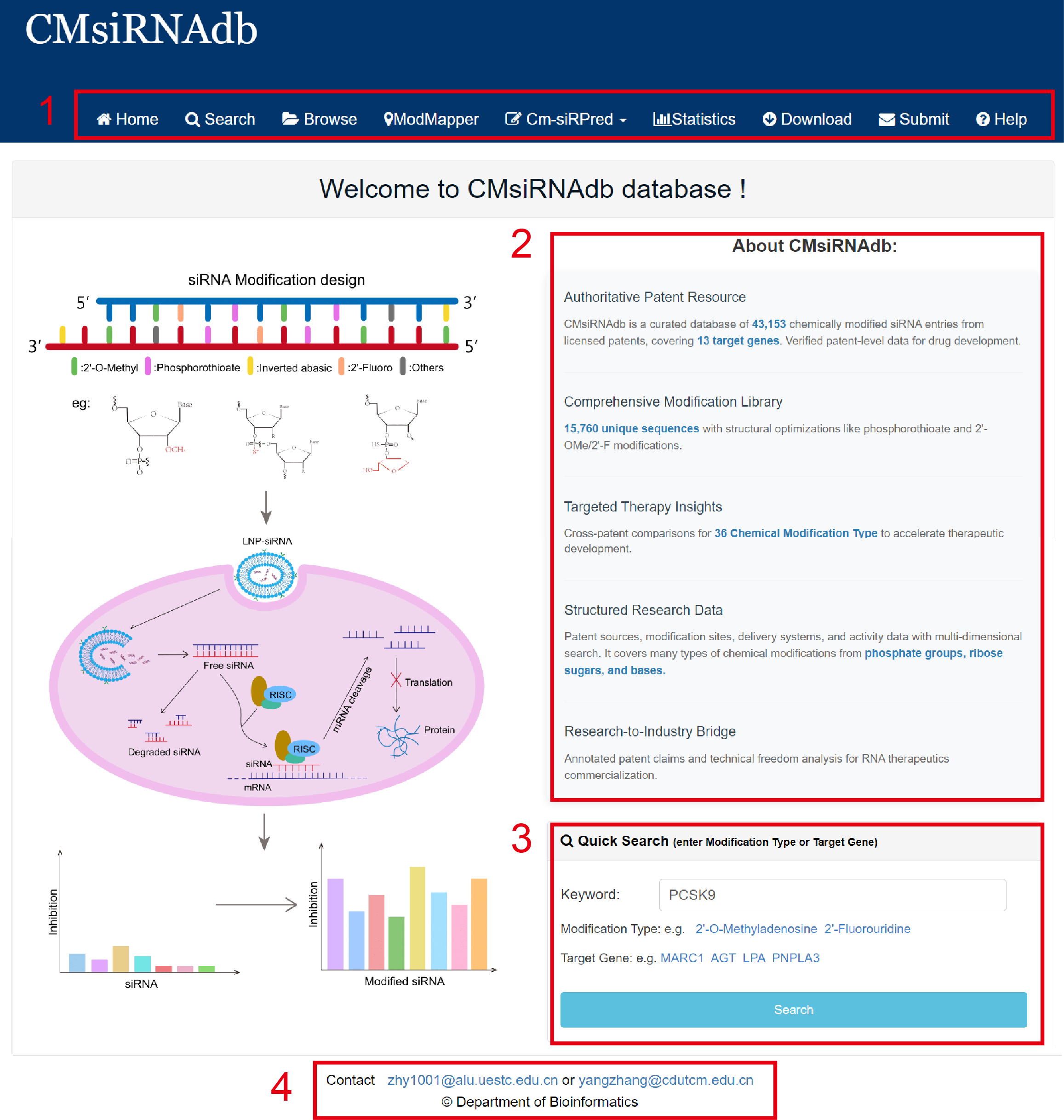
Fig 1-1. Home page
The search page is displayed in Fig 2-1, 2-2 and 2-3:
Exact search:
1. Select exact search.
2. Select type of input keyword: four choices are provided (Modifications in sense strand/Modifications in antisense strand/Position in Sense strand/Position in Antisense strand).
3. Input a keyword corresponding to selected type.For details about the type of modification, please refer to Question 7
4. Select the cell type to filter the search (39 cell types are available for reference).
5. Select target gene to filter the search (13 genes: PNPLA3, HSD17B13, AGT, LPA, INHBE, PCSK9, APP, MAPT, MARC1, CTNNB1, ANGPTL3, PLN, MSTN).
6. Select Modification locations to filter the search (Include touch-ups in different positions: Phosphate, Base, Sugar/Phosphate, Base/Phosphate, Sugar/Phosphate/Base/Sugar).
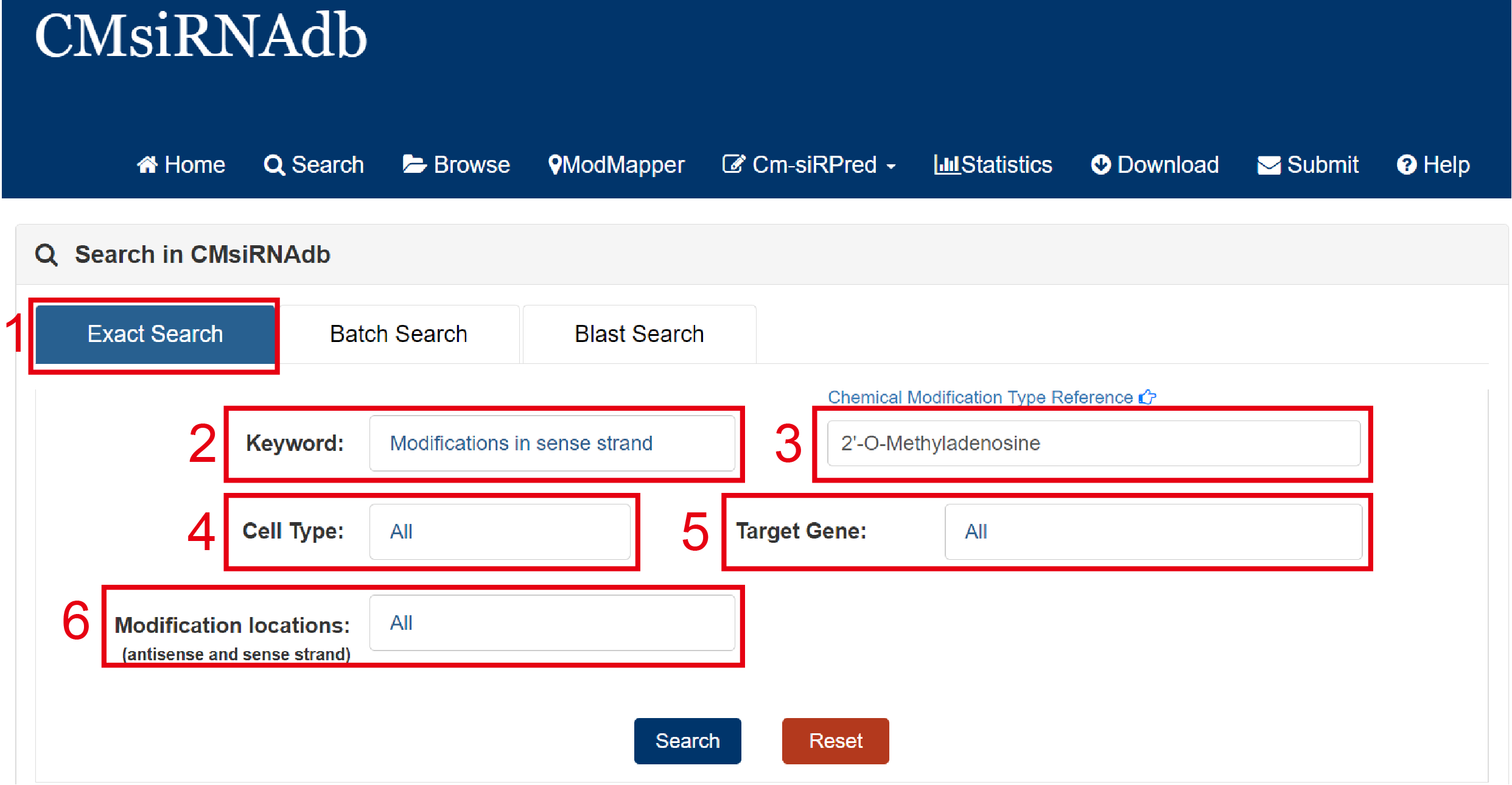
Fig 2-1. Exact Search page
Batch search:
1. Select batch search.
2. Select type of input keyword: three choices are provided (Modification Type/Target Gene/Patent ID).
3. Input list of keywords corresponding to selected type.
4. Select the cell type to filter the search (39 cell types are available for reference).
5. Select Modification locations to filter the search (Include touch-ups in different positions: Phosphate, Base, Sugar).
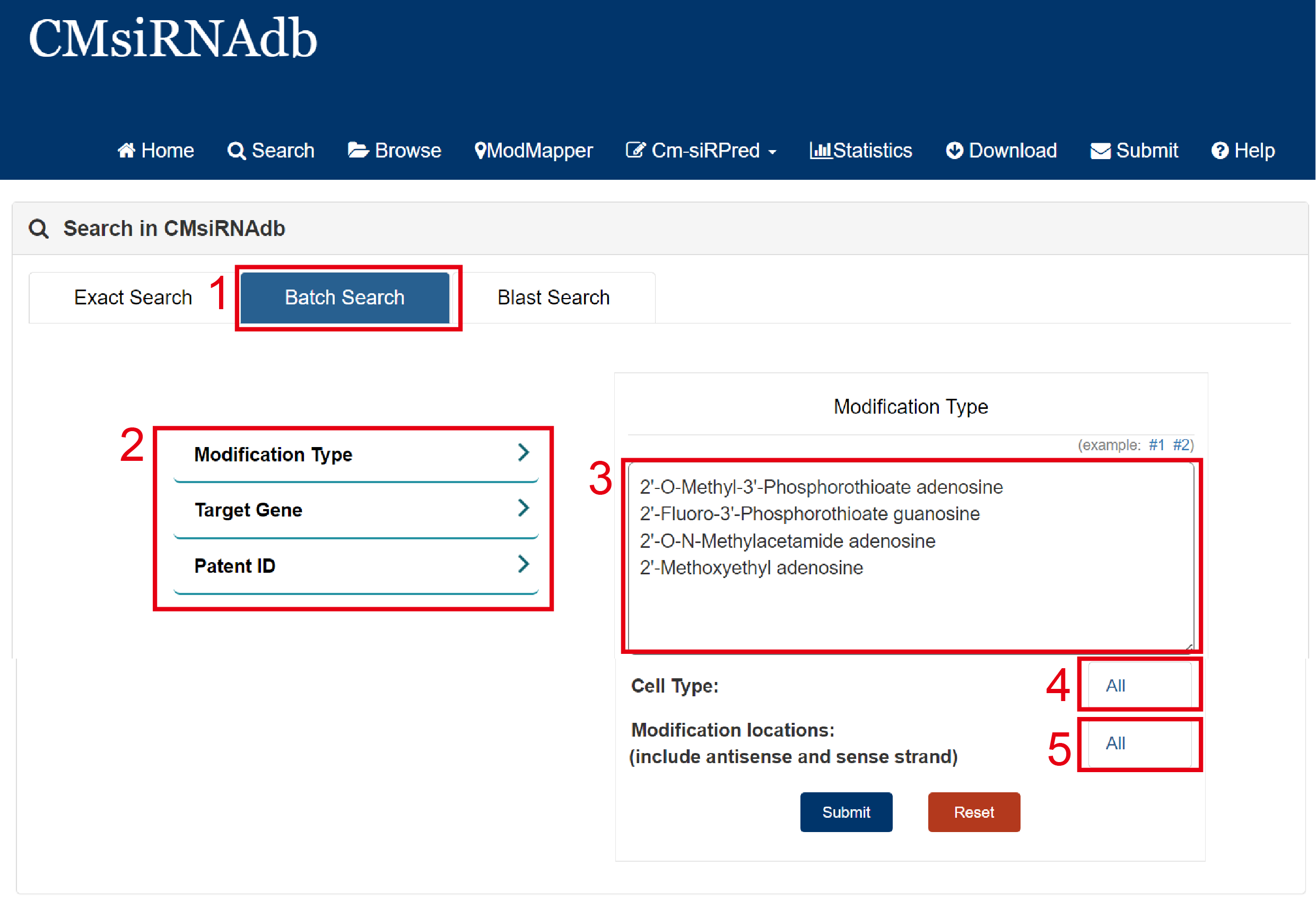
Fig 2-2. Batch Search page
Blast search:
1. Select Blast search.
2. Enter the sequence information in FASTA format in the text box.
3. Click example to view an example.
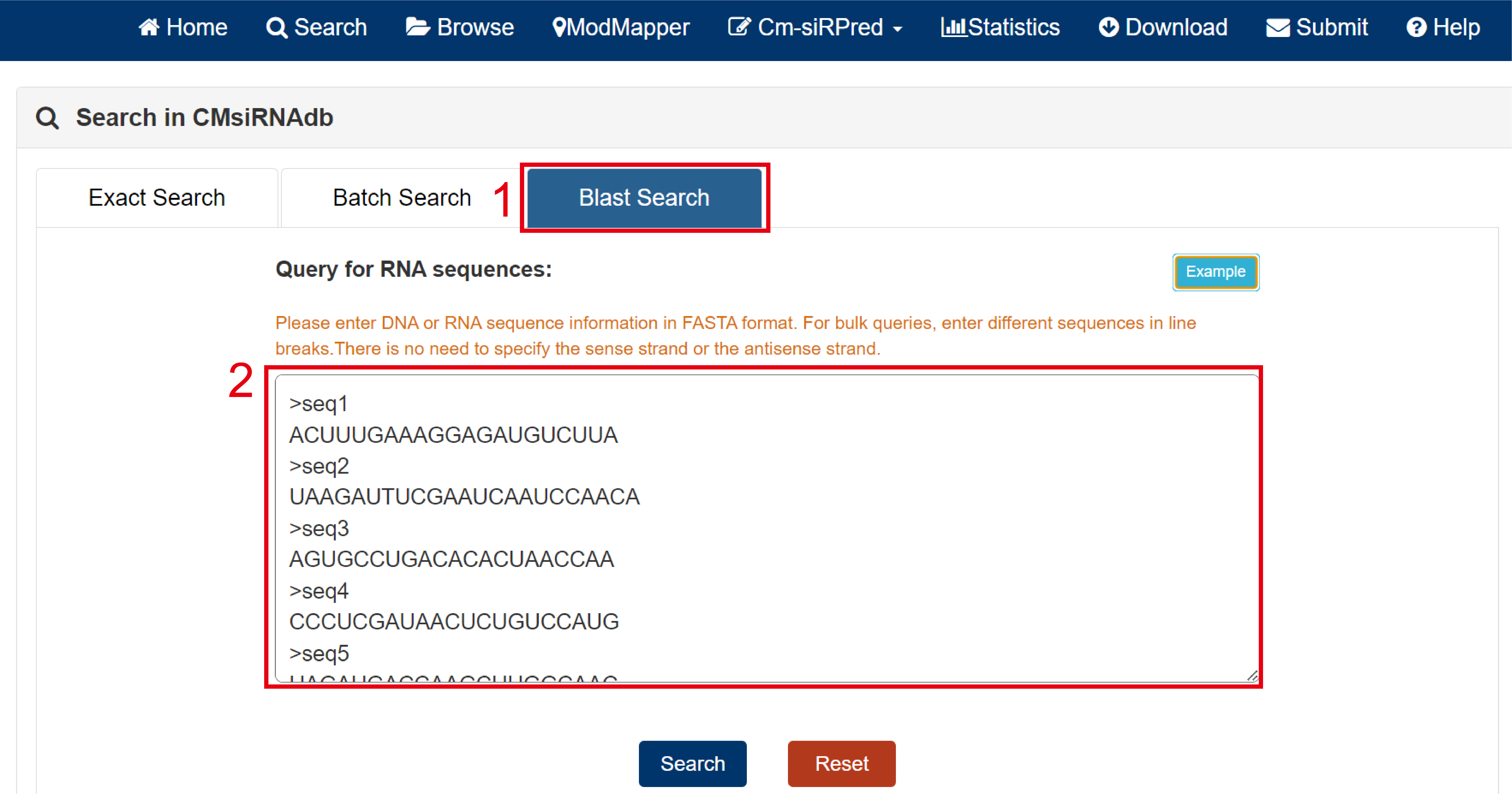
Fig 2-3. Blast Search page
Result page of Exact search and Batch search:
For the result page of Exact search and Batch search, all entries are listed with basic information including Patent ID, Target Gene, Cell Type, Modification sequence information(include sense strands and antisense strands), inhibition rate. Click more for more details
Fig 3-1:
1. Search keyword from the result table.
2. The result table (including Patent ID, Target Gene, Cell Type).
3. Click to link to Detail page.
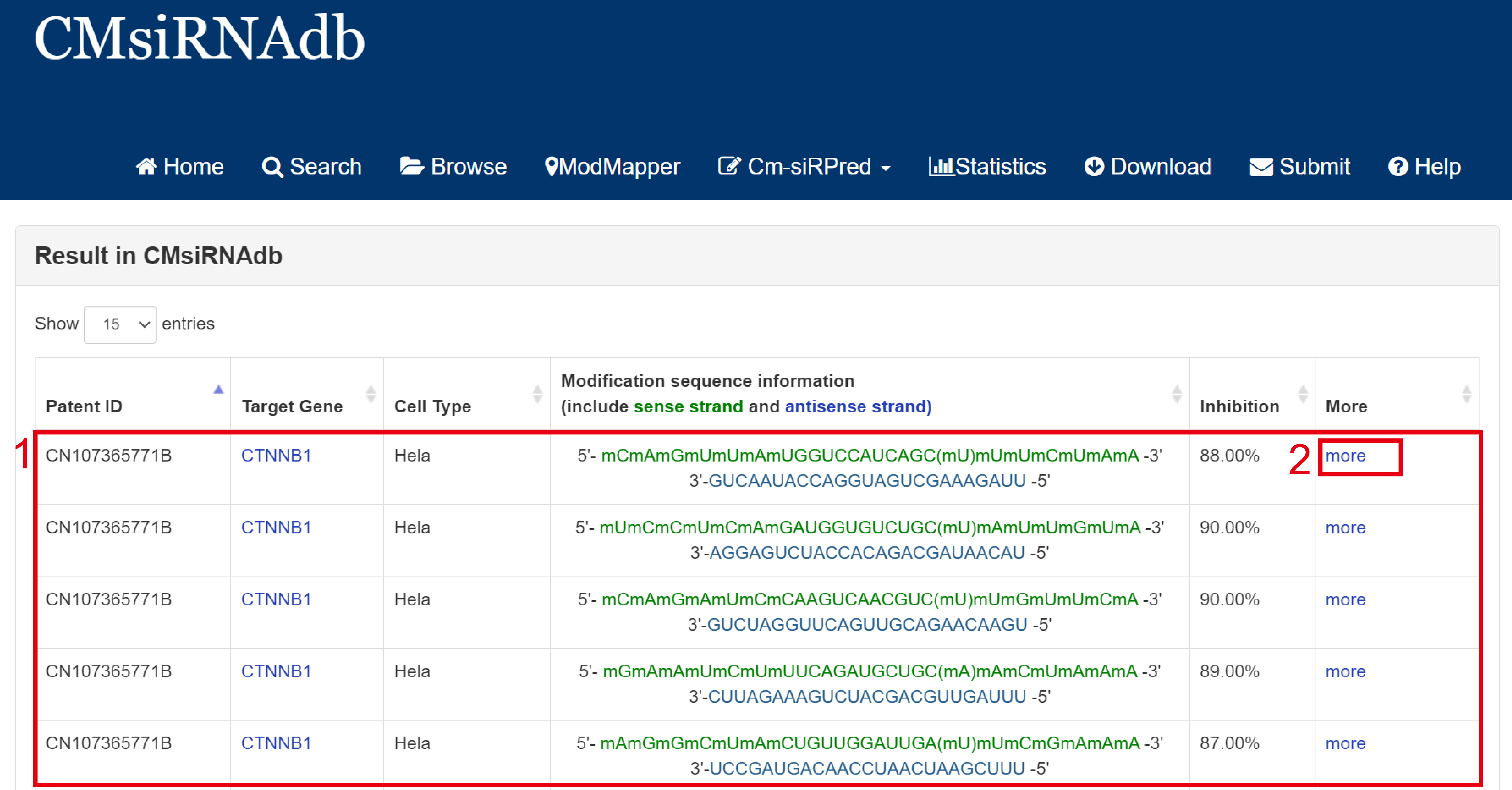
Fig 3-1. Result page of Exact search and Batch search
Result page of Blast search:
For the result page of Blast search, all entries are listed with basic information including Patent ID, Target Gene, Cell Type, Modification sequence information(include sense strands and antisense strands), inhibition rate, start and end of the matched sequence of documented proteins and Bit-score.
Fig 3-2:
1. Search keyword from the result table.
2. The result table (including Patent ID, Target Gene, Cell Type).
3. Click to link to Detail page.
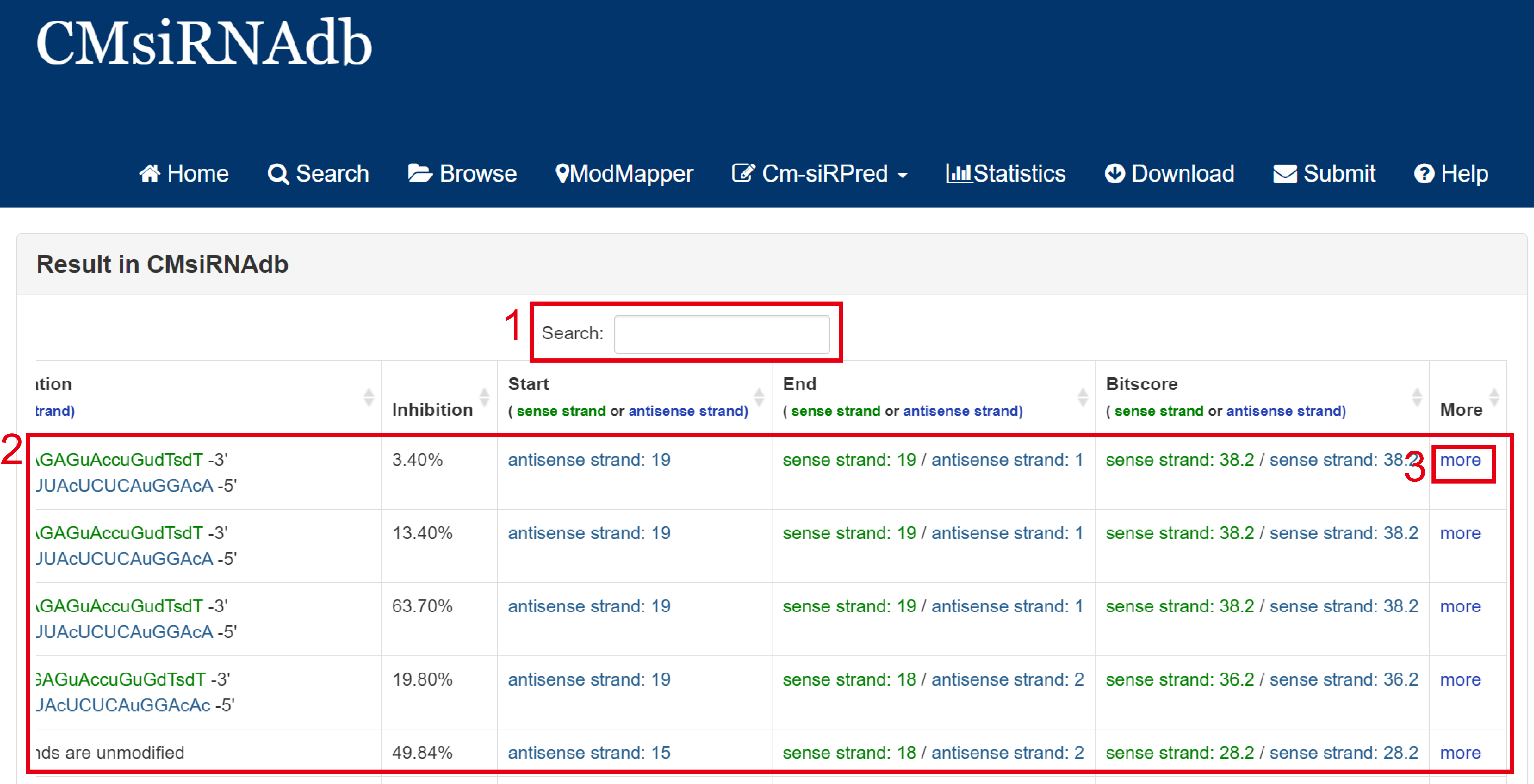
Fig 3-2. Result page of Blast search
On the details page, you can view various details of the data, which are displayed in three categories: Patent Basic Information, Sequence Information, Chemical Modification Mapping Table.
Fig 4-1:
1. Patent Basic Information: including Patent ID, Authorization status, Accession number, Patent Title(Non-English titles are marked with []), Target Gene, Gene ID, Cell Type, Concentration, Time_of_administration, Inhibition and Standard deviation.
2. Sequence Information: including Raw Sense/Antisense strand, Chemically modification seuqence, sequence length, modification annotation information, Modification Component and Modification positions. The above information includes both the sense strands and the antisense strands
3. Chemical Modification Mapping Table: the Modification, Description and Category of the modification. The types of modifications present in the current sequence will be further annotated in the table
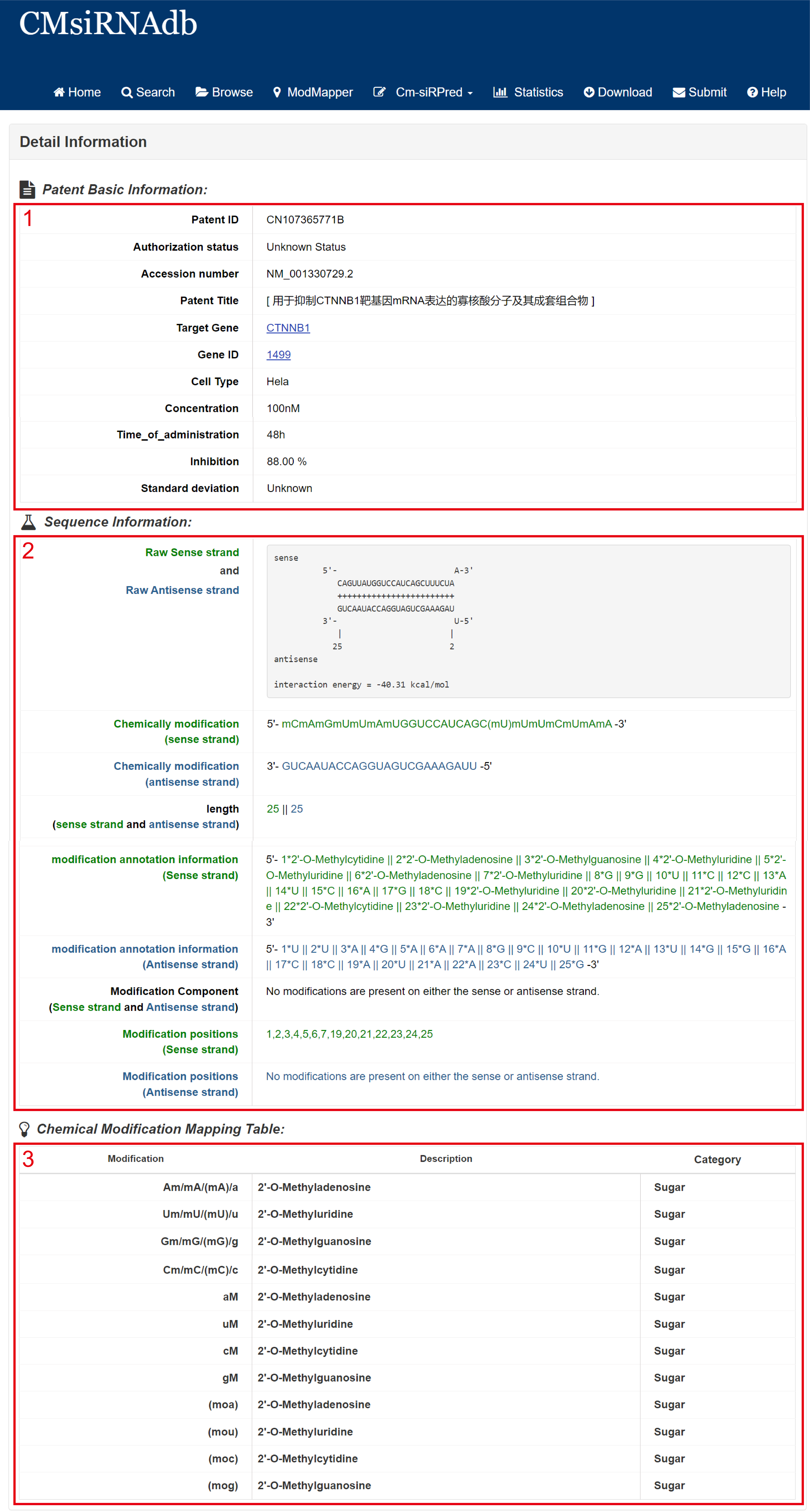
Fig 4-1. Detail page
The siRNA sequence information was presented in the Browse page, Users can browse all the information by three filter ways: by Cell Type, by Modification Type and by Modification location.Selecting an option will filter based on the data that has already been displayed and return the data that meets the requirements
Fig 5-1:
1. Filter by Cell Type (39 cell types are available for reference).
2. Filter by Modification Type (A variety of combined chemical modification types are available).
3. Filter by Modification location (Include touch-ups in different positions: Phosphate, Base, Sugar).
4. The result table.

Fig 5-1. Browse page
To empower researchers in deciphering nucleotide-specific chemical modification patterns, we developed ModMapper - an
intelligent sequence analysis tool. This system employs advanced parsing algorithms to automatically deconstruct
modification units at single-nucleotide resolution, providing:
1. Precise identification of chemical modification types
2. Cleavage position mapping
The interactive visualization interface enables rapid localization of target modifications. ModMapper delivers molecular-level insights critical for rational
siRNA therapeutic design, significantly enhancing modification pattern analysis efficiency.
Fig 6-1:
1. Enter the original sequence information and the chemical modification sequence information in FASTA format in the text box on the right.
2. If there are rare types of groomes that require special attention, please fill them in the third text box, or you may not fill them out.
3. Click to Submit and wait for results.Batch analysis is supported.

Fig 6-1. ModMapper page
Result page of ModMapper:
On the ModMapper analysis results page, we split the input chemical modification sequences by modification unit, gave detailed chemical modification annotation information on each nucleotide, and counted the sequence length, and for non-nucleotide modifications, we represented them as target groups.
Fig 6-2:
1. Position: The position of the nucleotide of this modifying unit in the original sequence.
2. Original sequence unit: Modification units in the input protochemical modification sequence.
3. Modification: Chemical modification annotation information.
4. Location: The localization of this chemical modification on a base or phosphate group or ribose.
5. Download All: Download all sequence information for analysis.
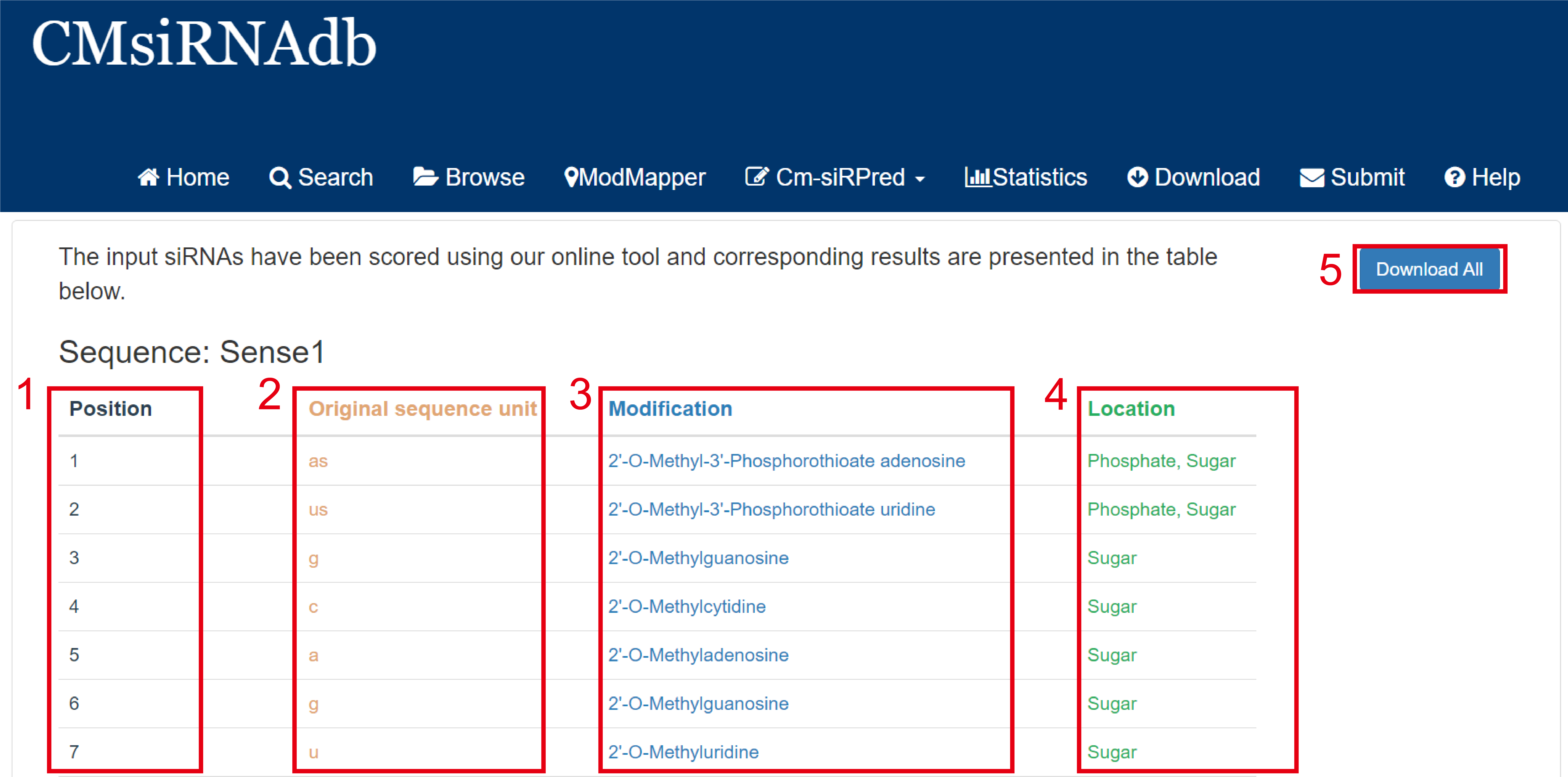
Fig 6-2. Results page of ModMapper
About Cm-siRPred:
Cm-siRPred is a specialized platform for cm-siRNA drug development, offering two core modules:
1. Chemical Modification Design - Enables real-time dynamic design of chemical modifications
2. Efficiency Prediction Streamlines batch processing of cm-siRNA sequences (FASTA) and modification patterns (SMILES)
This integrated solution accelerates rational design of RNA-based therapeutics through precision modification
optimization and efficacy prediction capabilities.
Fig 7-1:
1. Hover your mouse over Cm-siRPred in the navigation bar to display drop-down boxes about the designer module and the prediction module, click Cm-siRPred to enter the introduction page of Cm-siRPred.
2. Click The chemical modification design moudle to enter the designer page of Cm-siRPred.
3. Click The cm-siRNA efficiency prediction moudle to enter prediction module page of Cm-siRPred.
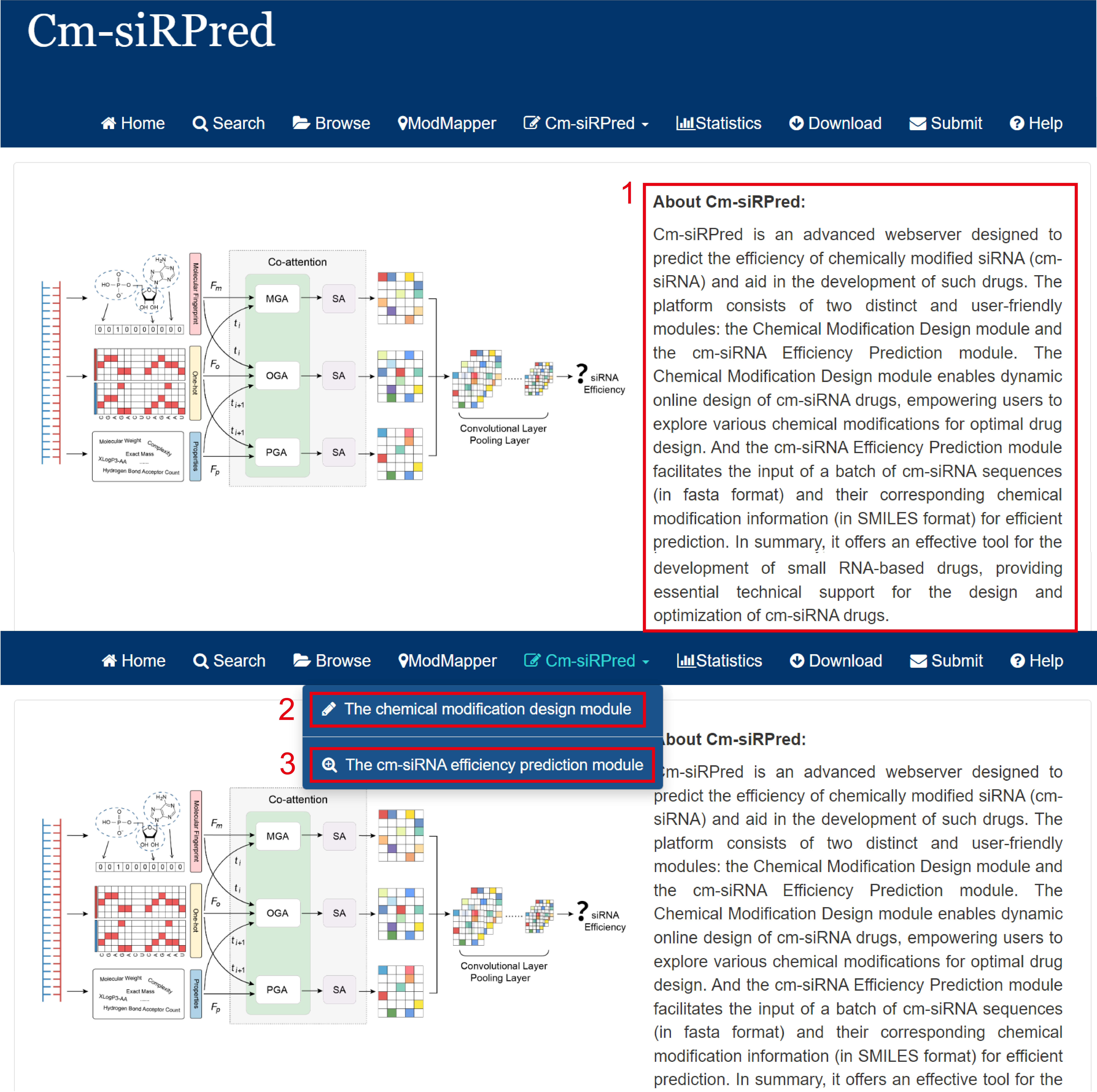
Fig 7-1. Cm-siRPred page
The designer module of Cm-siRPred
In the sequence design interface, you can select a unique chemical modification type for each base on the sense and antisense strands, support multiple chemical modification types on the same nucleotide, and our system will evaluate the inhibition efficiency score of this chemical modification combination after submission.
Fig 7-2:
1. Fill in the sequence information in FASTA format in the first text box, including the sense strands and the antisense strands.
2. Select the type of modification on each nucleotide on the sense and antisense strands, with support for multiple choices.
3. Click Submit to Server for analysis.
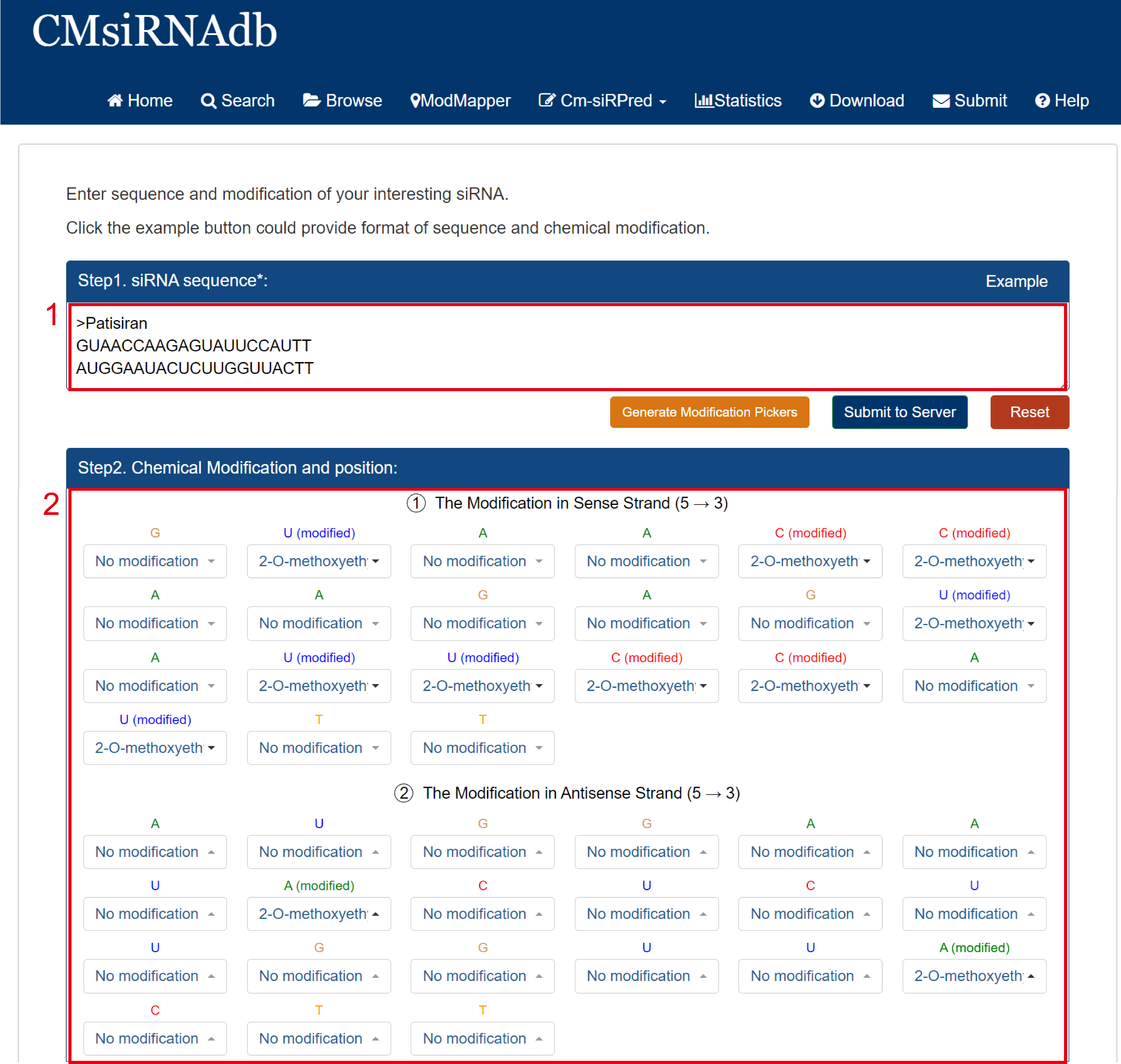
Fig 7-2. designer page of Cm-siRPred
The results page will give you the inhibition rate score for the combination of chemical modification types you selected.
Fig 7-3:
1. ID of siRNA: The name of the sequence that is specified when the sequence is entered.
2. Sequence of siRNA: siRNA sequence information.
3. Chemical Modification of siRNA: Information about the selected chemical modification sequence.
4. Score: Inhibition rate score.
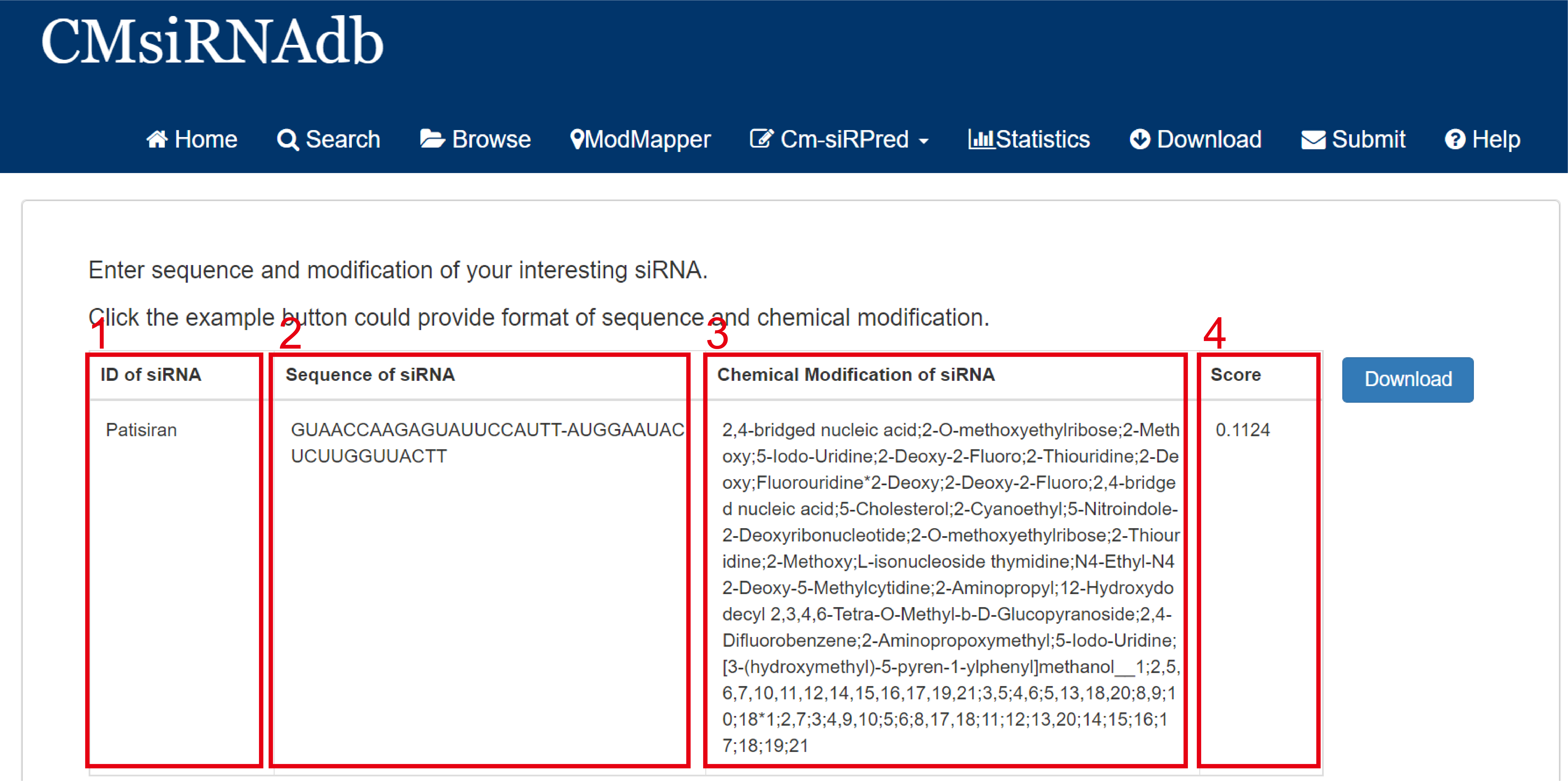
Fig 7-3. Analysis results of the designer module of Cm-siRPred
the prediction module of Cm-siRPred
Our prediction module of Cm-siRPred enables you to efficiently evaluate the chemical modification sequences you have already designed.
Fig 7-4:
1. Enter the original sequence information and the chemical modification sequence information in FASTA format in the text box on the right.
2. If there are rare types of groomes that require special attention, please fill them in the third text box, or you may not fill them out.
3. Click to Submit and wait for results.Batch analysis is supported.
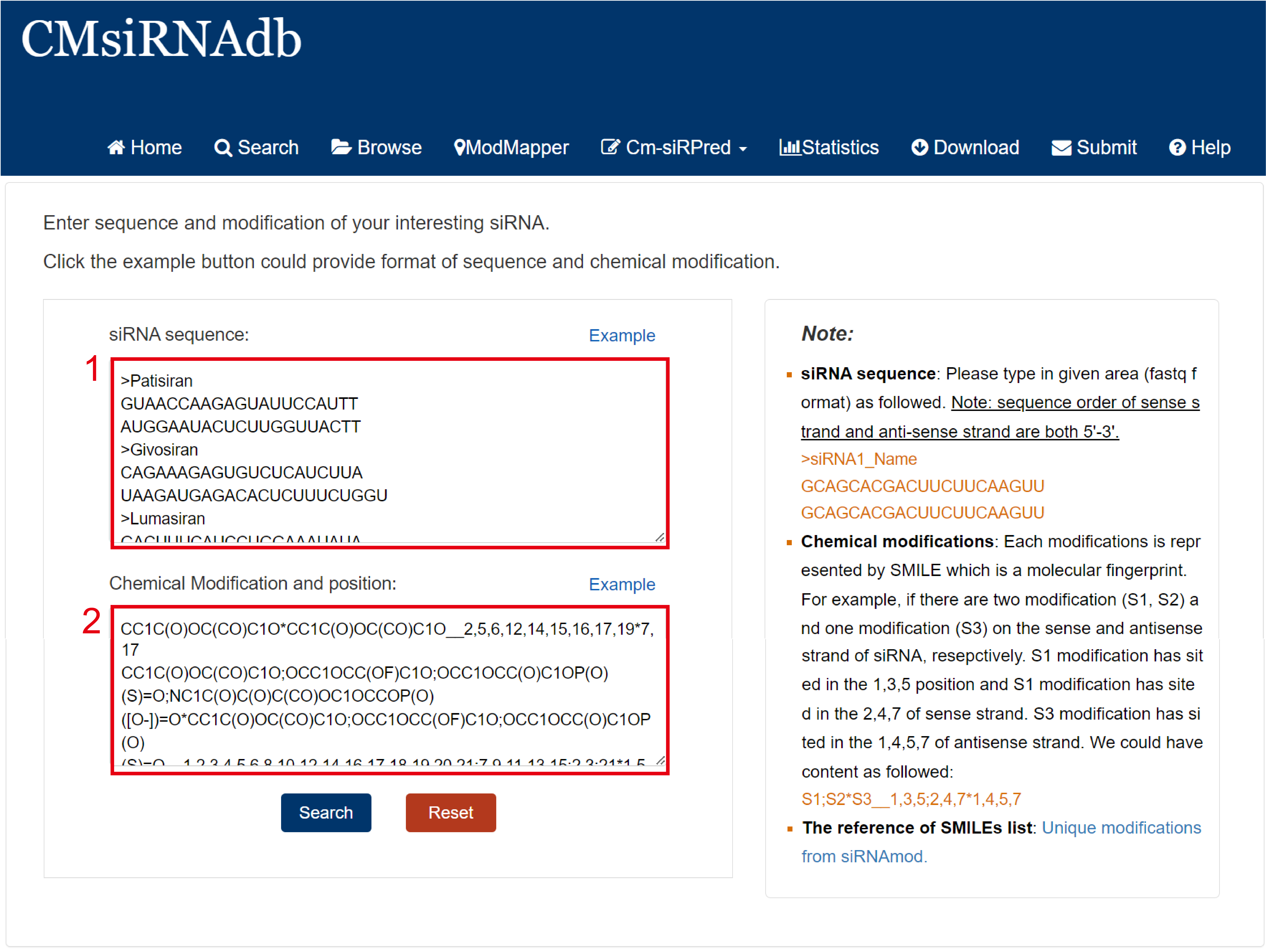
Fig 7-4. prediction module page of Cm-siRPred
The results page will give you the inhibition rate score for the combination of chemical modification types you submitted.
Fig 7-5:
1. ID of siRNA: The name of the sequence that is specified when the sequence is entered.
2. Sequence of siRNA: siRNA sequence information.
3. Chemical Modification of siRNA: Information about the selected chemical modification sequence.
4. Score: Inhibition rate score.
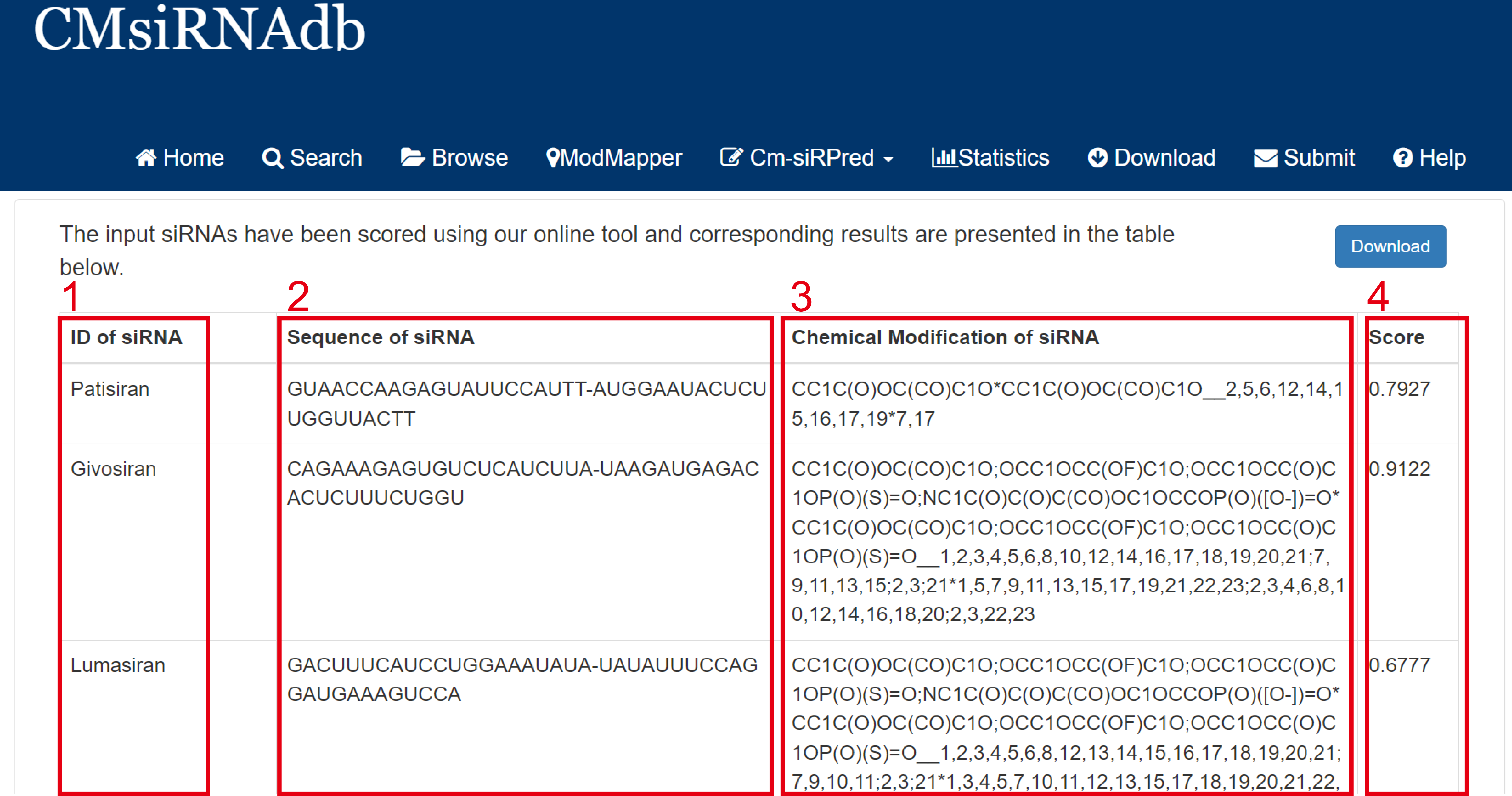
Fig 7-5. Analysis results of the prediction module of Cm-siRPred
CMsiRNAdb provide the download page for users. You can
download all the sequence data information in the collected patents in the download page.
There is also the option to download patent data that contains only a certain targeted gene
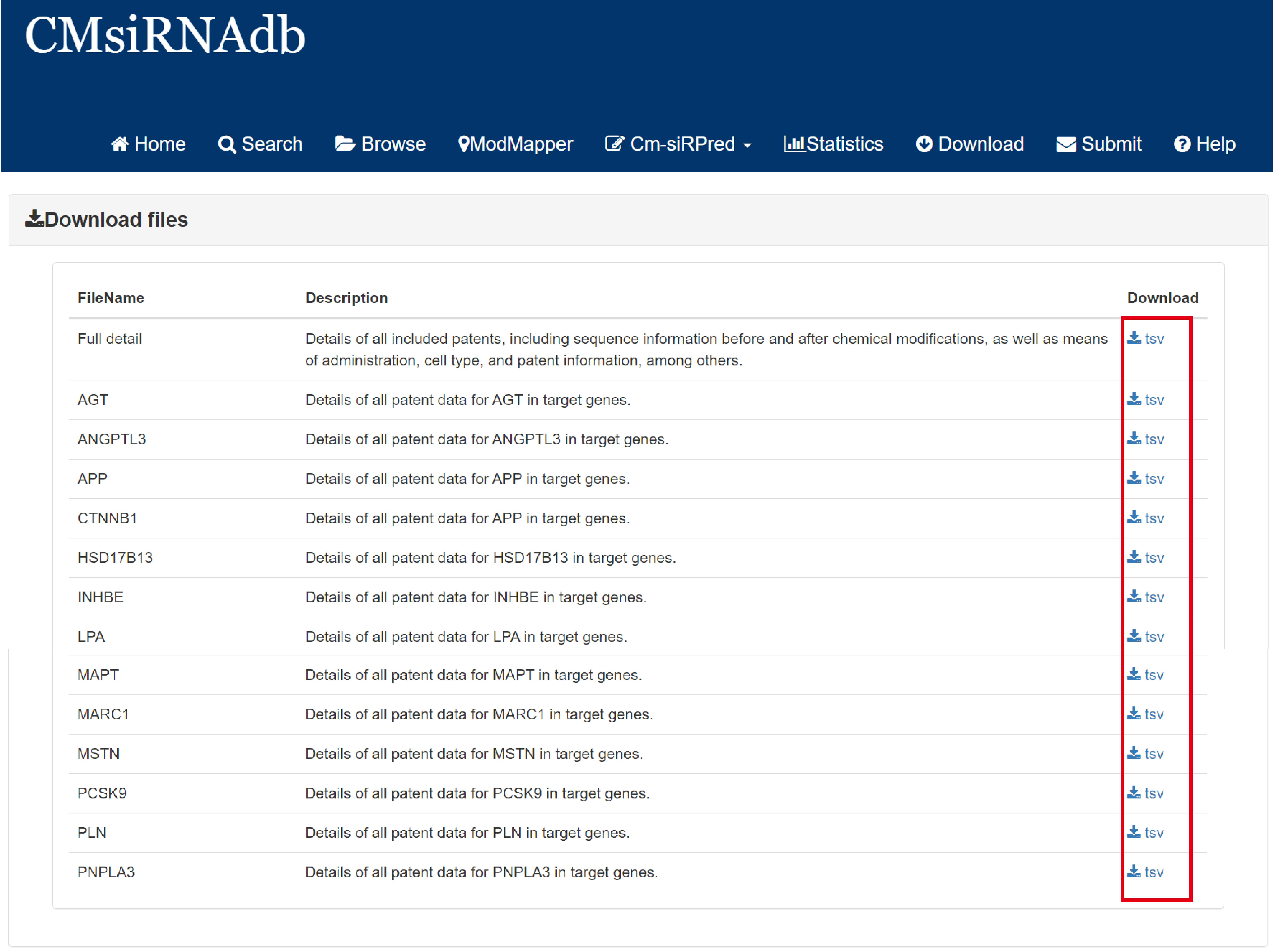
Fig 8-1. Download page
CMsiRNAdb provide specific chemical modification annotation information for the collected chemical modification sequences.
This is shown in the table below.
Chemical Modification Mapping Table:
| Modification | Description | Category |
|---|---|---|
| Am/mA/(mA)/a | 2'-O-Methyladenosine | Sugar |
| Ams/mAs | 2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| Gms/mGs | 2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| Gfs | 2'-Fluoro-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| Cms/mCs/mcs | 2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| Um/mU/(mU)/u | 2'-O-Methyluridine | Sugar |
| Ums/mUs | 2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| Gm/mG/(mG)/g | 2'-O-Methylguanosine | Sugar |
| Cm/mC/(mC)/c | 2'-O-Methylcytidine | Sugar |
| AmsEVP | 5'-Vinyl-(E)-phosphonate-2'-methoxyadenosine-3'-phosphorothioate | Sugar, Phosphate |
| UmsEVP | 5'-Vinyl-(E)-phosphonate-2'-methoxyuridine-3'-phosphorothioate | Sugar, Phosphate |
| Agna | Adenosine-glycol nucleic acid | Sugar |
| Cgna | Cytidine-glycol nucleic acid | Sugar |
| Ggna | Guanosine-glycol nucleic acid | Sugar |
| Tgna | Thymidine-glycol nucleic acid | Sugar |
| Ugna | Uridine-glycol nucleic acid | Sugar |
| (LNA-mC) | Locked Nucleic Acid in 2'-O-Methylcytidine | Sugar |
| (LNA-T) | Locked Nucleic Acid in Thymidine monophosphate | Sugar |
| (sLNA-T) | Phosphorothioate-Locked Nucleic Acid in Thymidine monophosphate | Sugar, Phosphate |
| (LNA-A) | Locked Nucleic Acid in Adenosine monophosphate | Sugar |
| (LNA-G) | Locked Nucleic Acid in Guanosine monophosphate | Sugar |
| (sLNA-G) | Phosphorothioate-Locked Nucleic Acid in Guanosine monophosphate | Sugar, Phosphate |
| (LNA-C) | Locked Nucleic Acid in Cytidine monophosphate | Sugar |
| (fA)/Af/fA | 2'-Fluoroadenosine | Sugar |
| (fU)/Uf/fU | 2'-Fluorouridine | Sugar |
| (fC)/Cf/fC | 2'-Fluorocytidine | Sugar |
| (fG)/Gf/fG | 2'-Fluoroguanosine | Sugar |
| dAs | 2'-Deoxy-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| dTs | 2'-Deoxy-3'-Phosphorothioate thymidine | Sugar, Phosphate |
| dCs | 2'-Deoxy-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| dG(s) | 2'-Deoxy-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| (invdT) | Reverse deoxythymidine | Sugar |
| (invdA) | Reverse deoxyadenosine | Sugar |
| Abs | Abasic-3'-Phosphorothioate | Sugar, Phosphate |
| Ab | Abasic | Sugar |
| AM | 2'-Methoxyethyl adenosine | Sugar |
| CM | 2'-Methoxyethyl cytidine | Sugar |
| UM | 2'-Methoxyethyl uridine | Sugar |
| GM | 2'-Methoxyethyl guanosine | Sugar |
| TM | 2'-Methoxyethyl thymidine | Sugar |
| AMs | 2'-Methoxyethyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| CMs | 2'-Methoxyethyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| UMs | 2'-Methoxyethyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| GMs | 2'-Methoxyethyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| TMs | 2'-Methoxyethyl-3'-Phosphorothioate thymidine | Sugar, Phosphate |
| (invAb) | Inverted abasic | Sugar |
| (invAb)sc | Inverted abasic-2'-O-Methyl-5'-Phosphorothioate cytidine | Sugar, Phosphate |
| (invAb)sa | Inverted abasic-2'-O-Methyl-5'-Phosphorothioate adenosine | Sugar, Phosphate |
| AunA | Adenosine modified with Unlocked Nucleic Acid | Sugar |
| CunA | Cytidine modified with Unlocked Nucleic Acid | Sugar |
| GunA | Guanosine modified with Unlocked Nucleic Acid | Sugar |
| UunA | Uridine modified with Unlocked Nucleic Acid | Sugar |
| (GNA-T) | Ethylene Glycol Nucleic Acid Thymidine | Phosphate |
| (TNA-U) | (L)-α-threofuranosyl Uridine | Sugar |
| (MeO-I) | 2'-O-Methylinosine | Sugar |
| (sMeO-I) | 3'-Thiophosphate-2'-O-Methylinosine | Sugar, Phosphate |
| Ahd | 2'-O-hexadecyl adenosine | Sugar |
| Uhd | 2'-O-hexadecyl uridine | Sugar |
| Chd | 2'-O-hexadecyl cytidine | Sugar |
| Ghd | 2'-O-hexadecyl guanosine | Sugar |
| (Ahd) | 2'-O-hexadecyl adenosine | Sugar |
| (Uhd) | 2'-O-hexadecyl uridine | Sugar |
| (Chd) | 2'-O-hexadecyl cytidine | Sugar |
| (Ghd) | 2'-O-hexadecyl guanosine | Sugar |
| Aam | 2'-O-N-Methylacetamide adenosine | Sugar |
| Tam | 2'-O-N-Methylacetamide thymidine | Sugar |
| Cam | 2'-O-N-Methylacetamide cytidine | Sugar |
| Gam | 2'-O-N-Methylacetamide guanosine | Sugar |
| (-)hmpNA(A) | (R)-2'-deoxy-2'-fluoro-4'-thioadenosine | Sugar |
| (-)hmpNA(U) | (R)-2'-deoxy-2'-fluoro-4'-thio-5-methyluridine | Sugar |
| (-)hmpNA(G) | (R)-2'-deoxy-2'-fluoro-4'-thioguanosine | Sugar |
| (-)hmpNA(C) | (R)-2'-deoxy-2'-fluoro-4'-thiocytidine | Sugar |
| mA*S | 2'-O-Methyl-3'-Phosphorothioate in the Sp configuration adenosine | Sugar, Phosphate |
| mU*S | 2'-O-Methyl-3'-Phosphorothioate in the Sp configuration uridine | Sugar, Phosphate |
| mC*S | 2'-O-Methyl-3'-Phosphorothioate in the Sp configuration cytidine | Sugar, Phosphate |
| mG*S | 2'-O-Methyl-3'-Phosphorothioate in the Sp configuration guanosine | Sugar, Phosphate |
| mA*R | 2'-O-Methyl-3'-Phosphorothioate in the Rp configuration adenosine | Sugar, Phosphate |
| mU*R | 2'-O-Methyl-3'-Phosphorothioate in the Rp configuration uridine | Sugar, Phosphate |
| mC*R | 2'-O-Methyl-3'-Phosphorothioate in the Rp configuration cytidine | Sugar, Phosphate |
| mG*R | 2'-O-Methyl-3'-Phosphorothioate in the Rp configuration guanosine | Sugar, Phosphate |
| fA*S | 2'-Fluoro-3'-Phosphorothioate in the Sp configuration adenosine | Sugar, Phosphate |
| fU*S | 2'-Fluoro-3'-Phosphorothioate in the Sp configuration uridine | Sugar, Phosphate |
| fC*S | 2'-Fluoro-3'-Phosphorothioate in the Sp configuration cytidine | Sugar, Phosphate |
| fG*S | 2'-Fluoro-3'-Phosphorothioate in the Sp configuration guanosine | Sugar, Phosphate |
| fA*R | 2'-Fluoro-3'-Phosphorothioate in the Rp configuration adenosine | Sugar, Phosphate |
| fU*R | 2'-Fluoro-3'-Phosphorothioate in the Rp configuration uridine | Sugar, Phosphate |
| fC*R | 2'-Fluoro-3'-Phosphorothioate in the Rp configuration cytidine | Sugar, Phosphate |
| fG*R | 2'-Fluoro-3'-Phosphorothioate in the Rp configuration guanosine | Sugar, Phosphate |
| aF | 2'-Fluoroadenosine | Sugar |
| cF | 2'-Fluorocytidine | Sugar |
| uF | 2'-Fluorouridine | Sugar |
| gF | 2'-Fluoroguanosine | Sugar |
| aM | 2'-O-Methyladenosine | Sugar |
| uM | 2'-O-Methyluridine | Sugar |
| cM | 2'-O-Methylcytidine | Sugar |
| gM | 2'-O-Methylguanosine | Sugar |
| aF* | 2'-Fluoro-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| cF* | 2'-Fluoro-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| uF* | 2'-Fluoro-3'-Phosphorothioate uridine | Sugar, Phosphate |
| gF* | 2'-Fluoro-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| fAs | 2'-Fluoro-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| fCs | 2'-Fluoro-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| fUs | 2'-Fluoro-3'-Phosphorothioate uridine | Sugar, Phosphate |
| fGs | 2'-Fluoro-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| mA* | 2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| mU* | 2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| mC* | 2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| mG* | 2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| fA* | 2'-Fluoro-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| fC* | 2'-Fluoro-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| fU* | 2'-Fluoro-3'-Phosphorothioate uridine | Sugar, Phosphate |
| fG* | 2'-Fluoro-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| mA(s) | 2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| mU(s) | 2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| mC(s) | 2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| mG(s) | 2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| fAps | 2'-Fluoro-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| fCps | 2'-Fluoro-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| fUps | 2'-Fluoro-3'-Phosphorothioate uridine | Sugar, Phosphate |
| fGps | 2'-Fluoro-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| mAps | 2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| mUps | 2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| mCps | 2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| mGps | 2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| mA*dQ | Inverted abasic deoxyribose-2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| mU*dQ | Inverted abasic deoxyribose-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| mC*dQ | Inverted abasic deoxyribose-2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| mG*dQ | Inverted abasic deoxyribose-2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| (ccBPmU)ps | 5'-O-Cyclopentylcarbamoyl-Pseudouridine-5'-Monophosphate-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| As | 3'-Phosphorothioate adenosine | Phosphate |
| Cs | 3'-Phosphorothioate cytidine | Phosphate |
| Gs | 3'-Phosphorothioate guanosine | Phosphate |
| Us | 3'-Phosphorothioate uridine | Phosphate |
| Ts | 3'-Phosphorothioate thymidine | Phosphate, Base |
| p-a | 5'-Phosphate ribose-2'-O-Methyl adenosine | Sugar, Phosphate |
| p-c | 5'-Phosphate ribose-2'-O-Methyl cytidine | Sugar, Phosphate |
| p-g | 5'-Phosphate ribose-2'-O-Methyl guanosine | Sugar, Phosphate |
| p-u | 5'-Phosphate ribose-2'-O-Methyl uridine | Sugar, Phosphate |
| P-u | 5'-Phosphate ribose-2'-O-Methyl uridine | Sugar, Phosphate |
| P-Uf | 5'-Phosphate ribose-2'-Fluoro uridine | Sugar, Phosphate |
| cPrpTMs | Cyclopropyl Phosphonate-2'-Methoxyethyl-3'-Phosphorothioate thymidine | Sugar, Phosphate |
| cPrpus | Cyclopropyl Phosphonate-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| PlmUs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| PlmAs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| PlmCs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| PlmGs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| Plus | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| Plas | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| Plcs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| Plgs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| (Phos) | 5'-Phosphate ribose | Phosphate |
| (Phos)us | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| (Phos)as | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| (Phos)cs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| (Phos)gs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| (Phosphate)us | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| (Phosphate)as | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate adenosine | Sugar, Phosphate |
| (Phosphate)cs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate cytidine | Sugar, Phosphate |
| (Phosphate)gs | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate guanosine | Sugar, Phosphate |
| Pl-Ums | 5'-Phosphate ribose-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| Ps-Ums | Phosphorothioate-2'-O-Methyl-3'-Phosphorothioate uridine | Sugar, Phosphate |
| MePhosphonate-4O-mUs | 2'-O-methyl-4'-O-methylene phosphonate-uridine | Phosphate, Sugar |
| (moa) | 2'-O-Methyladenosine | Sugar |
| (mou) | 2'-O-Methyluridine | Sugar |
| (moc) | 2'-O-Methylcytidine | Sugar |
| (mog) | 2'-O-Methylguanosine | Sugar |
| Tmoe | 2'-Methoxyethyl thymidine | Sugar |
| Umoe | 2'-Methoxyethyl uridine | Sugar |
| Cmoe | 2'-Methoxyethyl cytidine | Sugar |
| Gmoe | 2'-Methoxyethyl guanosine | Sugar |
| Y1 | Y1 | |
| I | 2'-Methoxy hypoxanthine | Sugar, Base |
| (a2N) | 2'-O-Methyl-2'-aminoadenosine | Sugar, Base |
| (GLY-Ac) | Abasic | Sugar |
| (GLY-iBu) | Abasic | Sugar |
| ademA-GalNAc | 2'-O-methyladenosine-N-acetylgalactosamine | Sugar |
| (C6NH-L4U)/(C6NH-L15U)/(C6NH-L16U)/(C6NH-L17U)/(C6NH-L18U)/(C6NH-L19U)/(C6NH-L20U) | (C6NH-L4U)/(C6NH-L15U)/(C6NH-L16U)/(C6NH-L17U)/(C6NH-L18U)/(C6NH-L19U)/(C6NH-L20U) | Sugar |
| (J2-CONC16A)/(J2-CONC16U)/(J2-CONC16C) | (J2-CONC16adenosine)/(J2-CONC16uridine)/(J2-CONC16cytidine) | Sugar |
| H1*/H2*/H4*/H7*/H9*/Hd* | H1/H2/H4/H7/H9/Hd-Phosphorothioate | Phosphate |
| (GalNAc3K2AhxC6) | N-Acetylgalactosamine-triantennary-2-keto-2-azidohexanoic acid-C6 | Sugar |
| (sGalNAc3K2AhxC6) | Triantennary N-Acetylgalactosamine-triantennary-2-keto-2-azidohexanoic acid-C6 | Sugar |
| (sGalNAc3) | N-acetyl-galactosamine | Sugar |
| (GalNAc3) | N-acetyl-galactosamine | Sugar |
| GalNAc(L96) | N-[tris(GalNAc-alkyl)-amidodecanoyl)]-4-hydroxyprolinol(Hyp-(GalNAc-alkyl)3) | Sugar |
| -TriGalNAc | N-[tris(GalNAc-alkyl)-amidodecanoyl)]-4-hydroxyprolinol(Hyp-(GalNAc-alkyl)3) | Sugar |
| L96 | N-[tris(GalNAc-alkyl)-amidodecanoyl)]-4-hydroxyprolinol(Hyp-(GalNAc-alkyl)3) | Sugar |
| (C11-PEG3-NAG3) | C11-Polyethylene Glycol Tri-N-Acetylglucosamine | Sugar |
| (PAZ) | Piwi/Argonaute/Zwille domain | Phosphate |
| (C6-SS-Alk-Me) | C6-SS-Alkyl-Methyl | Sugar |
| (Chol-TEG) | Cholesterol-Triethylene Glycol | Phosphate |
| (Chol4) | Cholesterol-Triethylene Glycol | Phosphate |
| ps(Chol4) | 3'-Phosphorothioate-Cholesterol-Triethylene Glycol | Phosphate |
| NAG37s-IBs- | NAG37s-IBs | Phosphate |
| IB(-NAG0052') | IB(-NAG0052') | Phosphate |
| -IB | IB | Phosphate |
| (-NAG0052') | (-NAG0052') | Phosphate |
| L025L025 | Lipophilic Linker-025 Dimer | Phosphate |
| -LLL | L-shaped ligand | Phosphate |
| -SSS | S-shaped ligand | Phosphate |
| Mod001L001 | Mod001L001 | Phosphate |
| Y44 | 2'-Hydroxymethyl-Tetrahydrofuran-5'-Phosphate | Phosphate |
| z* | Targeting group-3'-Phosphorothioate | Phosphate |
| (GalNAc3)s | GalNAc3-Phosphorothioate | Phosphate |