Overview:
The current version of MCSdb documents about 7,000 manually curated MCS protein entries with experimental evidence, refers to more than 6000, 24 organelles and 44 MCSs across 11 species. And MCSdb also collected 263 protein complexes reside in different MCSs with experimental evidence. In summary, MCSdb provides a convenient and user-friendly interface for querying, browsing and visualizing detailed information about these MCS proteins, and will be of help in elucidating the functions and mechanisms of action of MCS proteins and promoting inter-organelle communication research.
The homepage is displayed in the following Fig 1-1:
1. Main functions of the database are provided in menu bar form (boxed in red).
2. Introduction and overview for MCSdb.
3. Quick search for users.

Fig 1-1. Home page
The search page is displayed in Fig 2-1, 2-2 and 2-3:
Exact search:
1. Select exact search.
2. Select type of input keyword: three choices are provided (Protein Symbol/Uniprot ID/Entrez ID).
3. Input a keyword corresponding to selected type.
4. Select the species to filter the search.
5. Select detected method to filter the search (3 methods: Low throughput experimental methods, Proximity Labeling Techniques and Mass-spectrometric techniques).
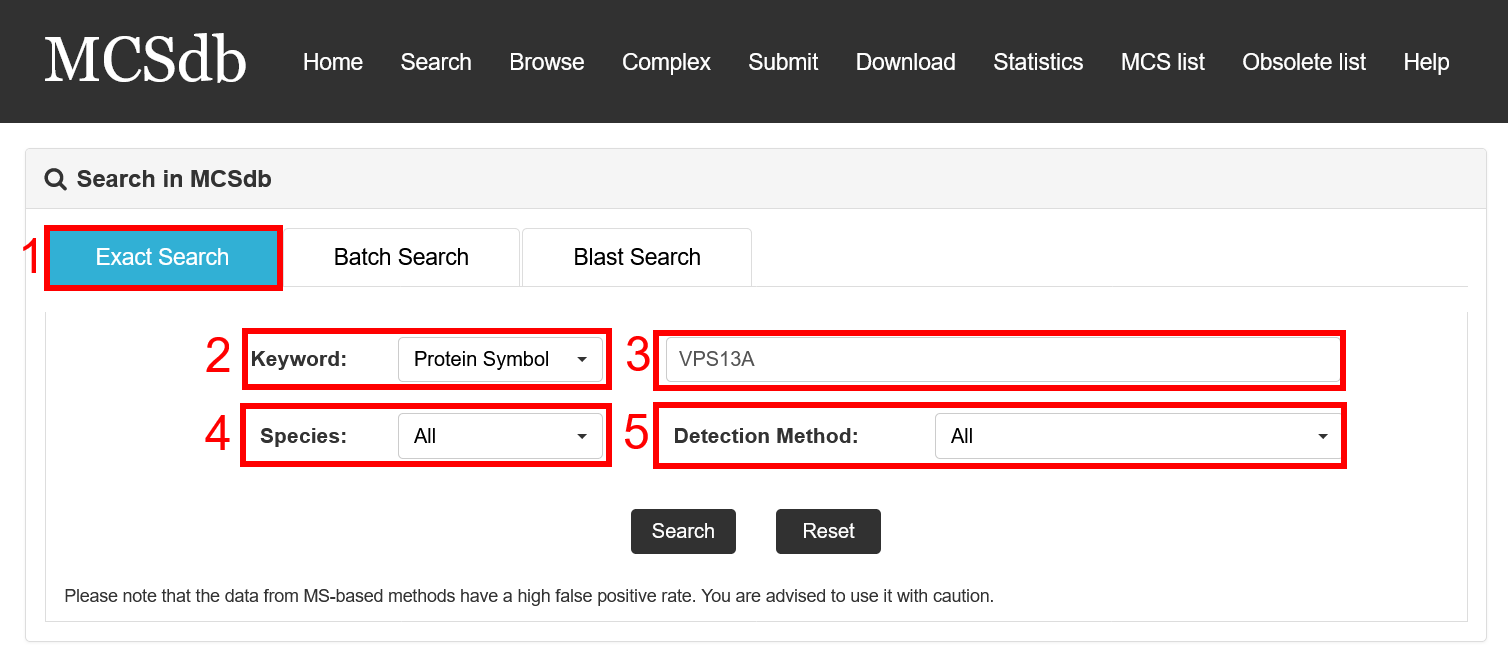
Fig 2-1. Exact Search page
Batch search:
1. Select batch search.
2. Select type of input keyword: three choices are provided (Protein Symbol/Uniprot ID/Entrez ID).
3. Input list of keywords corresponding to selected type.
4. Select the species to filter the search.
5. Select detected method to filter the search (3 methods: Low throughput experimental methods, Proximity Labeling Techniques and Mass-spectrometric techniques).
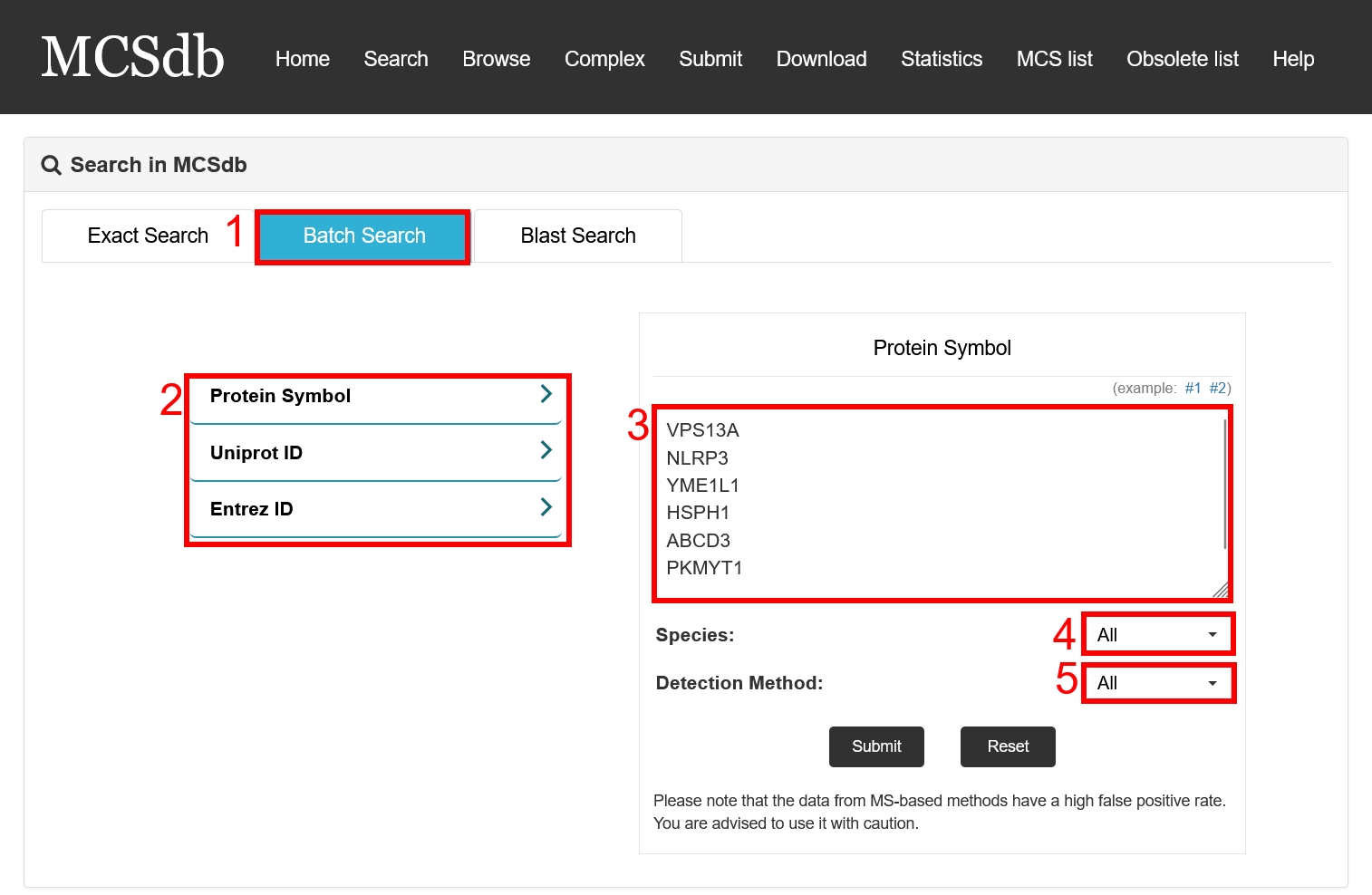
Fig 2-2. Batch Search page
Blast search:
1. Select Blast search.
2. Select type of input keyword: three choices are provided (Protein Symbol/Uniprot ID/Entrez ID).
3. Enter a sequence to do search.

Fig 2-3. Blast Search page
Result page of Exact search and Batch search:
For the result page of Exact search and Batch search, all entries are listed with basic information including gene ID, Uniprot ID, symbol, Species, MCS, Experimental method.
Fig 3-1:
1. Search keyword from the result table.
2. The result table (including gene ID, Uniprot ID, symbol, Species, MCS, Experimental method).
3. Click to link to Detail page.
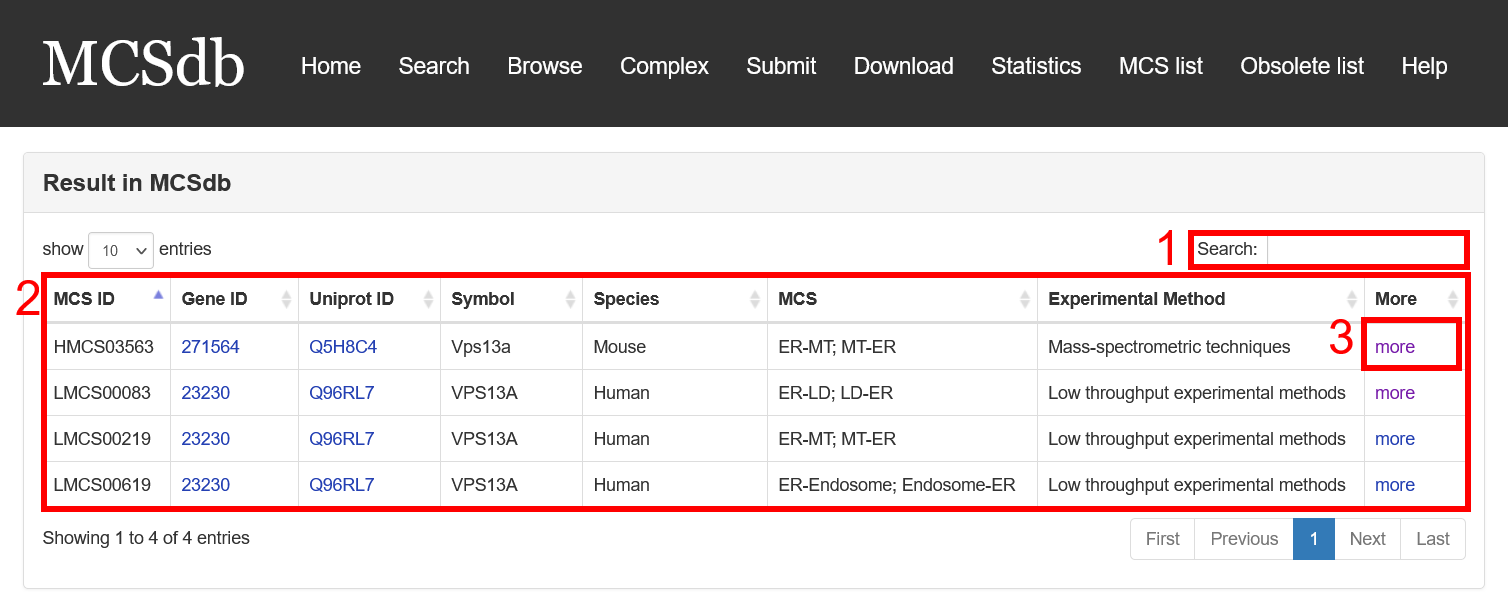
Fig 3-1. Result page of Exact search and Batch search
Result page of Blast search:
For the result page of Blast search, all entries are listed with basic information including gene ID, Uniprot ID, symbol, Species, MCS, Experimental method, start and end of the matched sequence of documented proteins and Bit-score.
Fig 3-2:
1. Search keyword from the result table.
2. The result table (including gene ID, Uniprot ID, symbol, Species, MCS, Experimental method).
3. Click to link to Detail page.
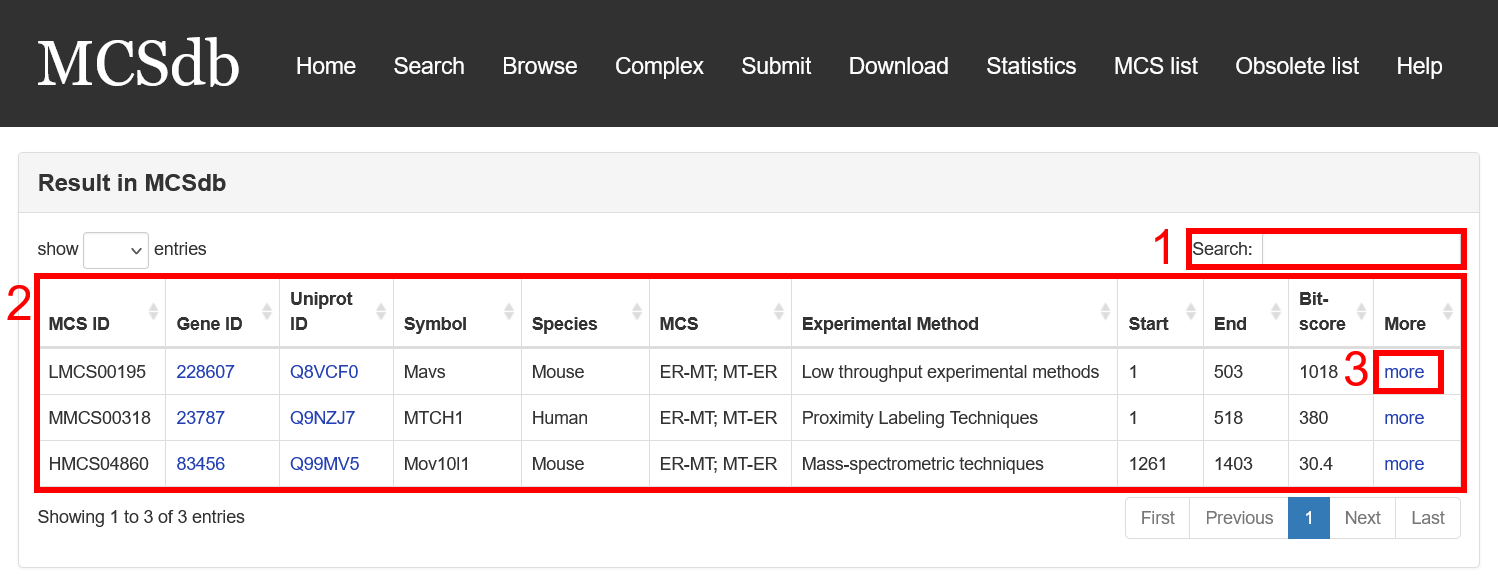
Fig 3-2. Result page of Blast search
In the Detail page, you can get the detail information of the MCS proteins including “Basic Information”, “Complex information”, “The expression of protein across different tissues”, “Homology Information (EggNOG, HOGENOM, OrthoDB, TreeFam and GeneTree databases)” and “References”.
Fig 4-1:
1. Basic Information: including Symbol, Species, Gene ID, Uniprot ID, Membrane Contact Site, Location (from literature), Cell line/Tissue, Experimental Method and protein sequence and protein sequence.
2. Complex information: including Complex ID, Subunit of complex, Subcellular location, Species and hyperlink to the detail page of complex.
3. The expression of MCS protein across different tissues.
4. Homology Information: including ortholog ID from EggNOG, HOGENOM, OrthoDB, TreeFam and GeneTree databases.
5. References: the PMID and description from literatures related to the MCS proteins.

Fig 4-1. Detail page
The MCS protein list was presented in the Browse page, Users can browse all the proteins by three filter ways: by species, by MCS and by detected method.
Fig 5-1:
1. Filter by species (11 type of species).
2. Filter by MCS (44 type of MCSs).
3. Filter by detected method (3 methods: Low throughput experimental methods, Proximity Labeling Techniques and Mass-spectrometric techniques).
4. The result table.
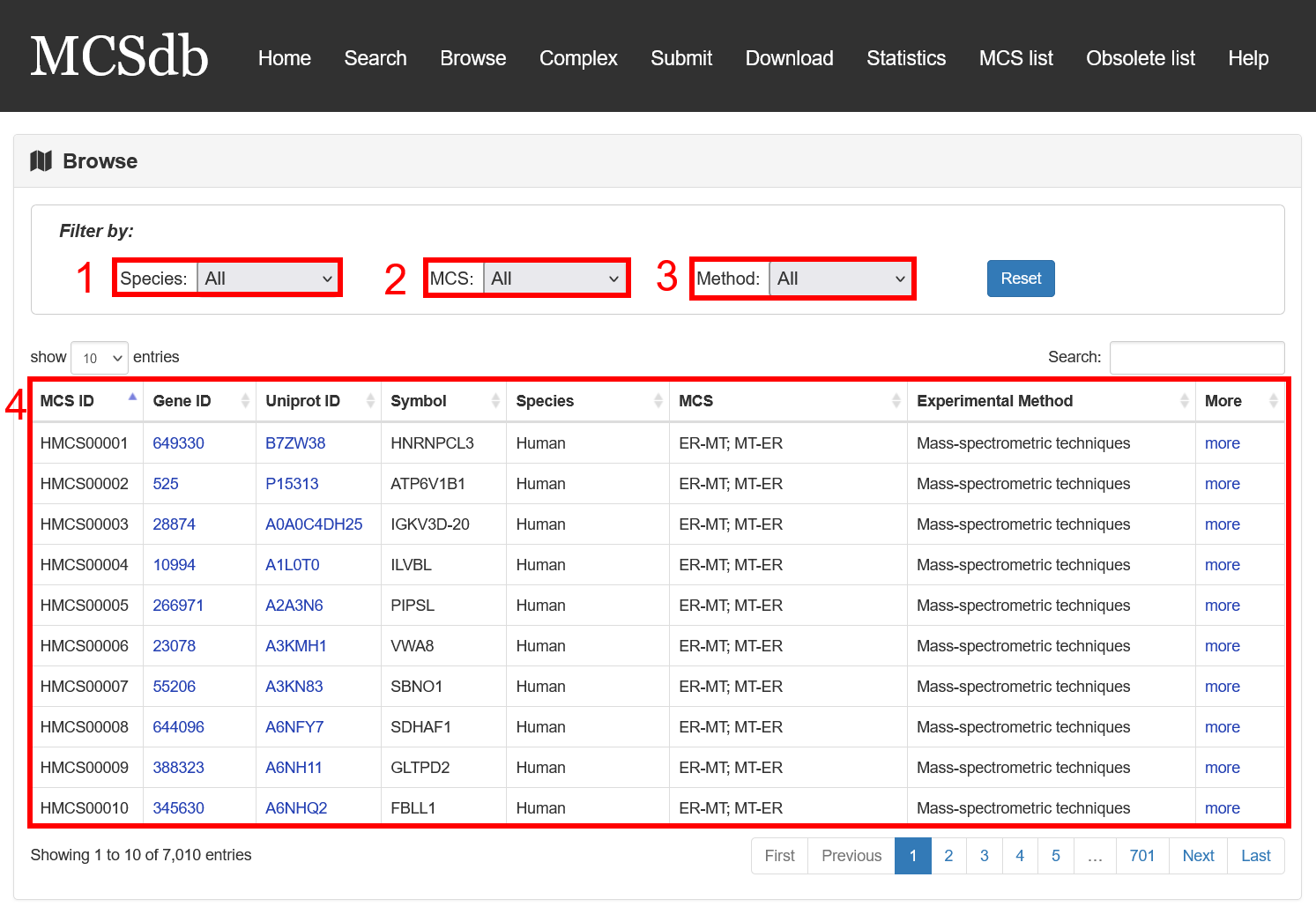
Fig 5-1. Browse page
To help the users to browse the complexes reside in MCS, MCSdb provides an independent webpage to query, browse and visualize detailed information about the 263 MCS complexes.
Fig 6-1:
1. Search keyword form the complex table.
2. Complex table (complex ID, subunit number, complex name, species and MCS).
3. Click to link to Detail page.

Fig 6-1. Complex page
In the Detail page of complex, you can get the detail information of the MCS complex including “Basic Information”, “Subunit information” and “References”.
Fig 6-2:
1. Basic Information: including Complex ID, Complex name, subunit number, Membrane Contact Site and species.
2. Subunit information: including subunit symbol, Uniprot ID, Subcellular location and hyperlink to the detail page of complex.
3. References: the PMID and description from literatures related to the MCS complex.
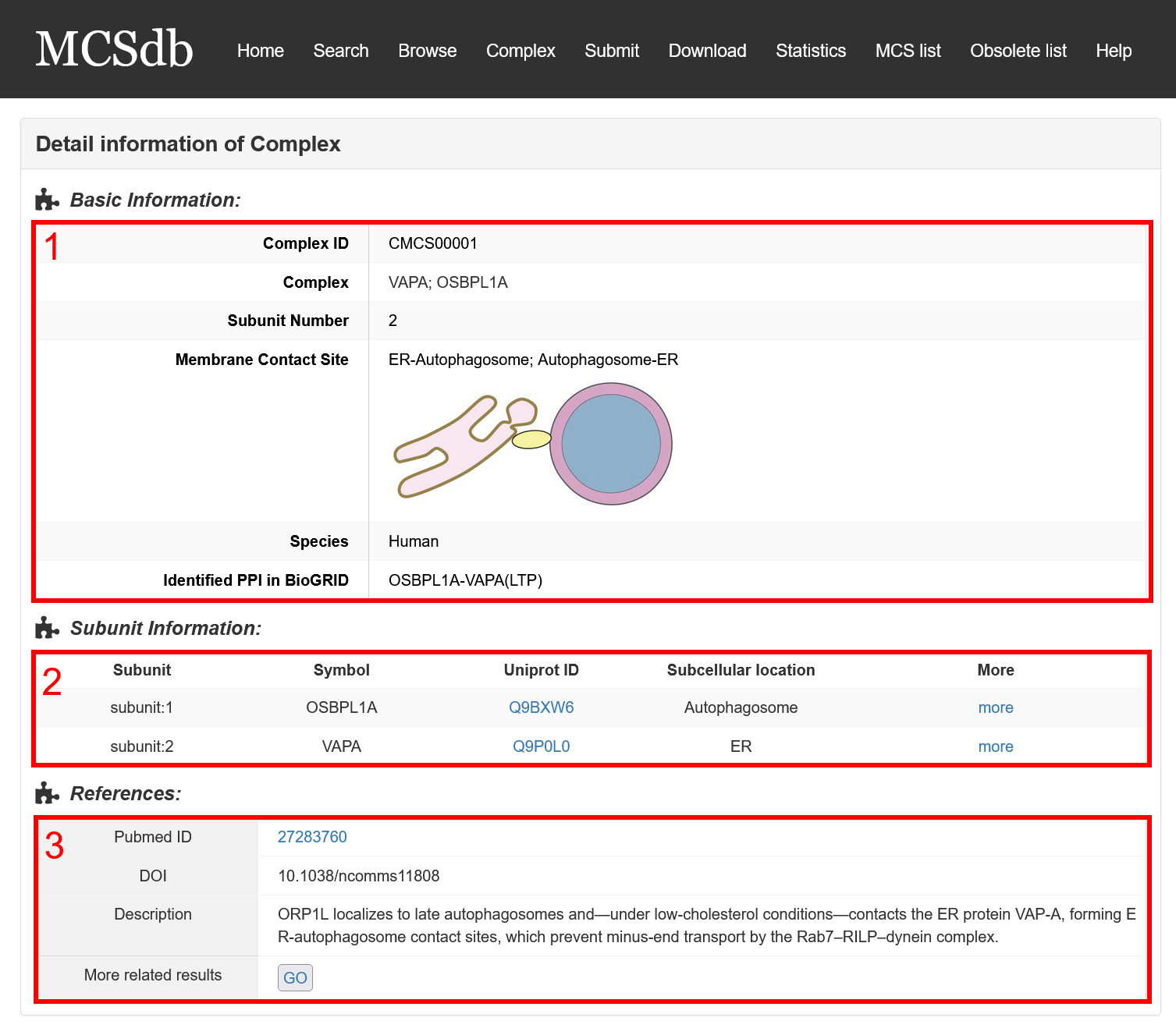
Fig 6-2. Detail page of complex
MCSdb provide the download page for users. You can download all the proteins and complexes data in the download page.
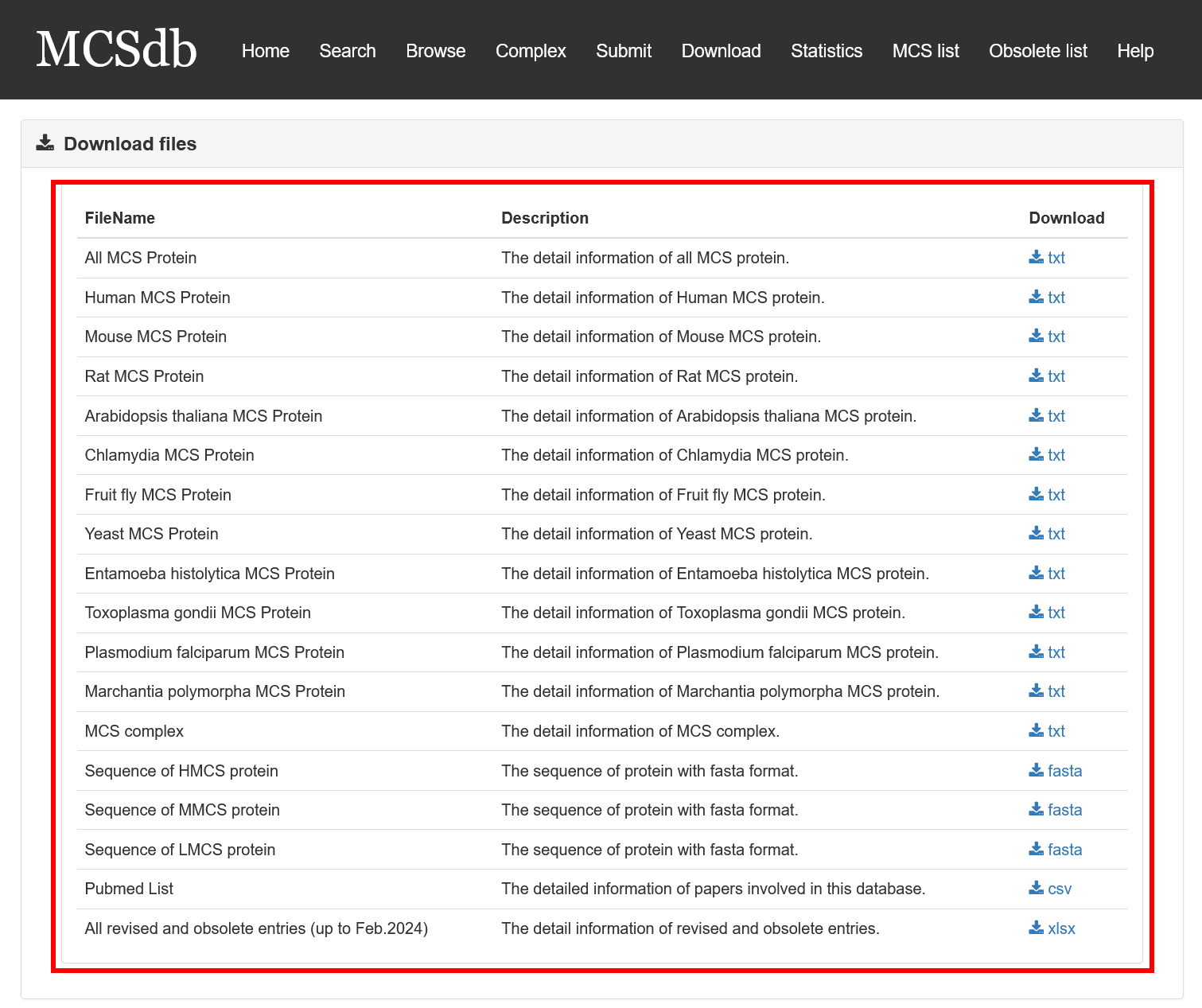
Fig 7-1. Download page
Throughout the process of curating data, we continually discovered mistakes and worked to improve. To prevent you from facing similar issues in your research, we've categorized the errors we've encountered into the following three categories. We share these insights for mutual enlightenment. For specific error instances, please refer to "All revised and obsolete entries (up to Feb.2024).xlsx ":
1. Misunderstanding the MCS Concept: This led to the erroneous inclusion of numerous proteins related to organelle budding, fission, and fusion (See Table S1 for relevant error instances).
MCSdb's Data Collection Standard for MCS:
Based on several review articles, the MCS is defined as an area of close apposition (ranging from 10 to 80 nm) between two bi- or mono-layer membrane-bound organelles. These organelles are physically connected via proteinaceous tethers but do not undergo fusion. As Scorrano, L. et al described in 2019, there are four essential features of MCS: (1) Tethering, (2) Lack of fusion, (3) Specific function, and (4) Defined proteome/lipidome. For a protein to be recognized as an MCS protein in MCSdb, there must be experimental evidence supporting its location at the MCS, or proof showing it can be attracted to the MCS, aiding its formation or functions related to the MCS.
2. Limitations from Team Constraints: Despite our best efforts, the limitations in our team's resources and scope of thought mean that our database may not capture every MCS protein (refer to table S2 for specifics). Therefore, we've introduced a submission interface, encouraging researchers to contribute information on MCS proteins that aren't yet documented in MCSdb. If you come across any MCS protein not listed in our database, we hope you to provide as much detailed information as possible to ensure precise data incorporation. Your invaluable contributions to MCSdb are deeply appreciated.
3. Past Work Shortcomings: Previously, there were lapses in meticulousness and rigor during data reading and collection. Coupled with occasional deficiencies in expert knowledge, this resulted in capturing erroneous information. Issues arose, such as misnaming proteins, misidentifying protein species, or mislabeling protein MCS locations (See table S3 for instances). If you identify any inaccurate data within the current database, we earnestly hope you can relay these issues to us promptly, either through email or the submission page.
Thank you for your understanding and collaboration!
We have collected the MCS proteins and complexes involved in the process of autophagy. The current version documents 31 complexes and 84 distinct proteins, spanning 13 different membrane contact sites and representing 7 diverse species:
9.1 Group by MCS:
| Serial number | MCS | Counts |
|---|---|---|
| 1 | ER-MT; MT-ER | 6219 |
| 2 | ER-PM; PM-ER | 154 |
| 3 | ER-Peroxisome; Peroxisome-ER | 91 |
| 4 | ER-LD; LD-ER | 87 |
| 5 | MT-Peroxisome; Peroxisome-MT | 71 |
| 6 | ER-Endosome; Endosome-ER | 52 |
| 7 | Peroxisome-Vacuole; Vacuole-Peroxisome | 47 |
| 8 | LD-PM; PM-LD | 41 |
| 9 | GA-Peroxisome; Peroxisome-GA | 38 |
| 10 | ER-GA; GA-ER | 33 |
| 11 | ER-Vacuole; Vacuole-ER | 20 |
| 12 | ER-LY; LY-ER | 18 |
| 13 | MT-LD; LD-MT | 18 |
| 14 | ER-Parasitophorous vacuole; Parasitophorous vacuole-ER | 9 |
| 15 | MT-Vacuole; Vacuole-MT | 10 |
| 16 | Nucleus-Vacuole; Vacuole-Nucleus | 10 |
| 17 | LY-GA; GA-LY | 8 |
| 18 | ER-Autophagosome; Autophagosome-ER | 7 |
| 19 | ER-Phg; Phg-ER | 7 |
| 20 | MT-Endosome; Endosome-MT | 6 |
| 21 | IM-ER; ER-IM | 5 |
| 22 | MT-LY; LY-MT | 5 |
| 23 | Peroxisome-LD; LD-Peroxisome | 4 |
| 24 | LD-LD | 4 |
| 25 | ER-Autolysosome; Autolysosome-ER | 4 |
| 26 | LD-Parasitophorous Vacuole; Parasitophorous Vacuole-LD | 4 |
| 27 | MT-PM; PM-MT | 3 |
| 28 | GA-Endosome; Endosome-GA | 3 |
| 29 | LY-Peroxisome; Peroxisome-LY | 3 |
| 30 | ER-Phagophore; Phagophore-ER | 3 |
| 31 | LD-Vacuole; Vacuole-LD | 3 |
| 32 | ER-MT-PM; PM-MT-ER | 2 |
| 33 | LY-LD; LD-LY | 2 |
| 34 | ER-MVB; MVB-ER | 2 |
| 35 | PM-Peroxisome; Peroxisome-PM | 2 |
| 36 | Endosome-Mitosome; Mitosome-Endosome | 2 |
| 37 | Nucleus-MT; MT-Nucleus | 2 |
| 38 | PM-WP body; WP body-PM | 2 |
| 39 | ER-Vesicle; Vesicle-ER | 2 |
| 40 | MT-Pellicle; Pellicle-MT | 2 |
| 41 | ER-Prospore membrane; Prospore membrane-ER | 2 |
| 42 | MT-GA; GA-MT | 1 |
| 43 | ER-Transport vesicle; Transport vesicle-ER | 1 |
| 44 | MT-Endolysosomes; Endolysosomes-MT | 1 |
9.2 Group by Organelle:
| Serial number | Organelle | Full name | Count |
|---|---|---|---|
| 1 | ER | Endoplasmic Reticulum | 6882 |
| 2 | MT | Mitochondria | 6340 |
| 3 | Peroxisome | / | 256 |
| 4 | PM | Plasmalemma | 204 |
| 5 | LD | Lipid Droplet | 163 |
| 6 | Vacuole | / | 106 |
| 7 | GA | Golgi Apparatus | 83 |
| 8 | Endosome | / | 62 |
| 9 | LY | Lysosome | 41 |
| 10 | Parasitophorous vacuole | / | 15 |
| 11 | Nucleus | / | 12 |
| 12 | Autophagosome | / | 7 |
| 13 | Phg | Phagosome | 7 |
| 14 | IM | Isolation membrane | 5 |
| 15 | Autolysosome | / | 4 |
| 16 | Phagophore | / | 3 |
| 17 | Mitosome | / | 2 |
| 18 | MVB | Multivesicular Bodies | 2 |
| 19 | Pellicle | / | 2 |
| 20 | Vesicle | / | 2 |
| 21 | WP body | Weibel-Palade body | 2 |
| 22 | Endolysosomes | / | 1 |
| 23 | Transport vesicle | / | 1 |
| 24 | Prospore membrane | / | 1 |
In order to facilitate the assessment of the reliability of MS based data by users, we propose a scoring system based on protein subcellular localization and protein-protein interaction (PPI) networks. We obtained the interaction information for MS-based proteins from the STRING database, and then acquired subcellular localization data from the Uniprot database to determine whether MS-based proteins and their interacting proteins are located in the MCS organelle.
This system categorizes MS based data into five levels of confidence, ranked from high to low as L1>L2>L3>L4>L5, encompassing the following ten scenarios: (Figure 10-1)
Fig 10-1:
L1: MS-based proteins are simultaneously located in two organelles within the MCS.
L2-1: MS-based proteins are located in one organelle within the MCS, while interacting proteins are located in another organelle.
L2-2: MS-based proteins are located in one organelle within the MCS with no interacting proteins.
L3-1: MS-based proteins are located in one organelle within the MCS, with interacting proteins not located in any organelle.
L3-2: MS-based proteins are located in one organelle within the MCS, with interacting proteins located in the same organelle.
L3-3: MS-based proteins are located in one organelle within the MCS with no interacting proteins.
L4-1: MS-based proteins are not located in any organelle within the MCS, with interacting proteins located in one organelle.
L4-2: MS-based proteins are not located in any organelle within the MCS, with interacting proteins simultaneously located in two organelles.
L5-1: MS-based proteins are not located in any organelle within the MCS with no interacting proteins.
L5-2: Both MS-based proteins and interacting proteins are not located in any organelle within the MCS.
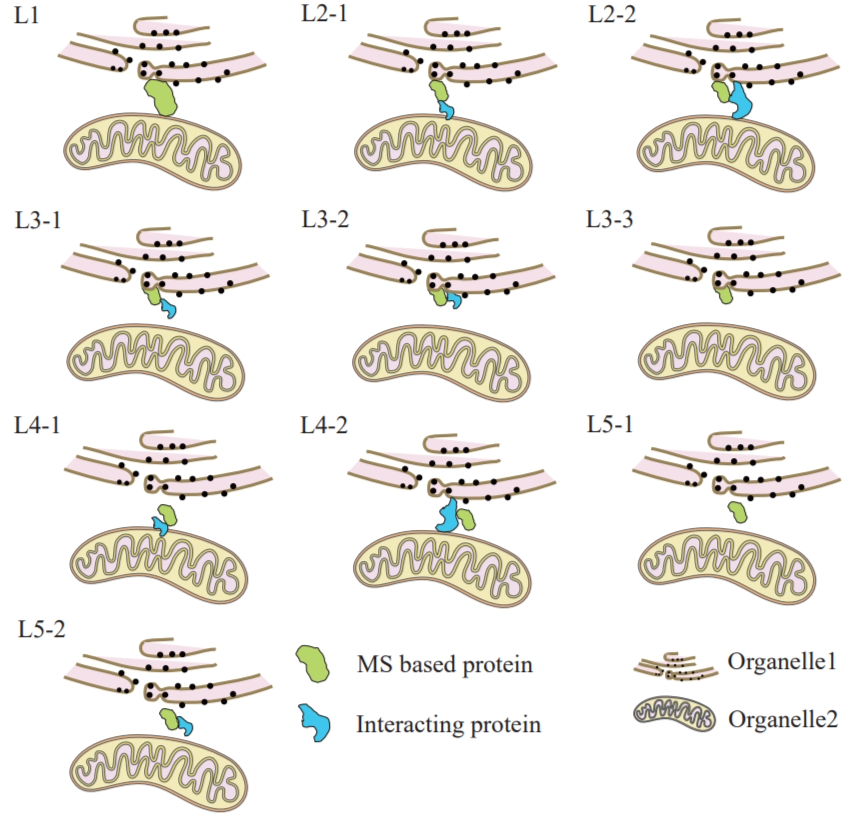
Fig 10-1 confidence score diagram
Fig 10-2:
In the set of 5985 mass-spectrometric data, the distribution of Confidence scores is shown in the following figure. (Figure 10-2)
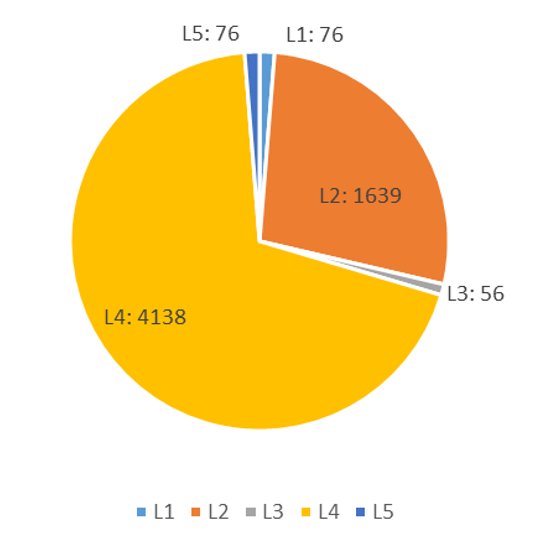
Fig 10-2 confidence score statistical graph
We have collected the MCS proteins and complexes involved in the process of autophagy. The current version documents 30 complexes and 84 distinct proteins, spanning 13 different membrane contact sites and representing 7 diverse species:
Map of autophagy-related MCS protein:
As shown in Figure 11-1, it's a circular diagram of Autophagy-related MCS proteins in our database.
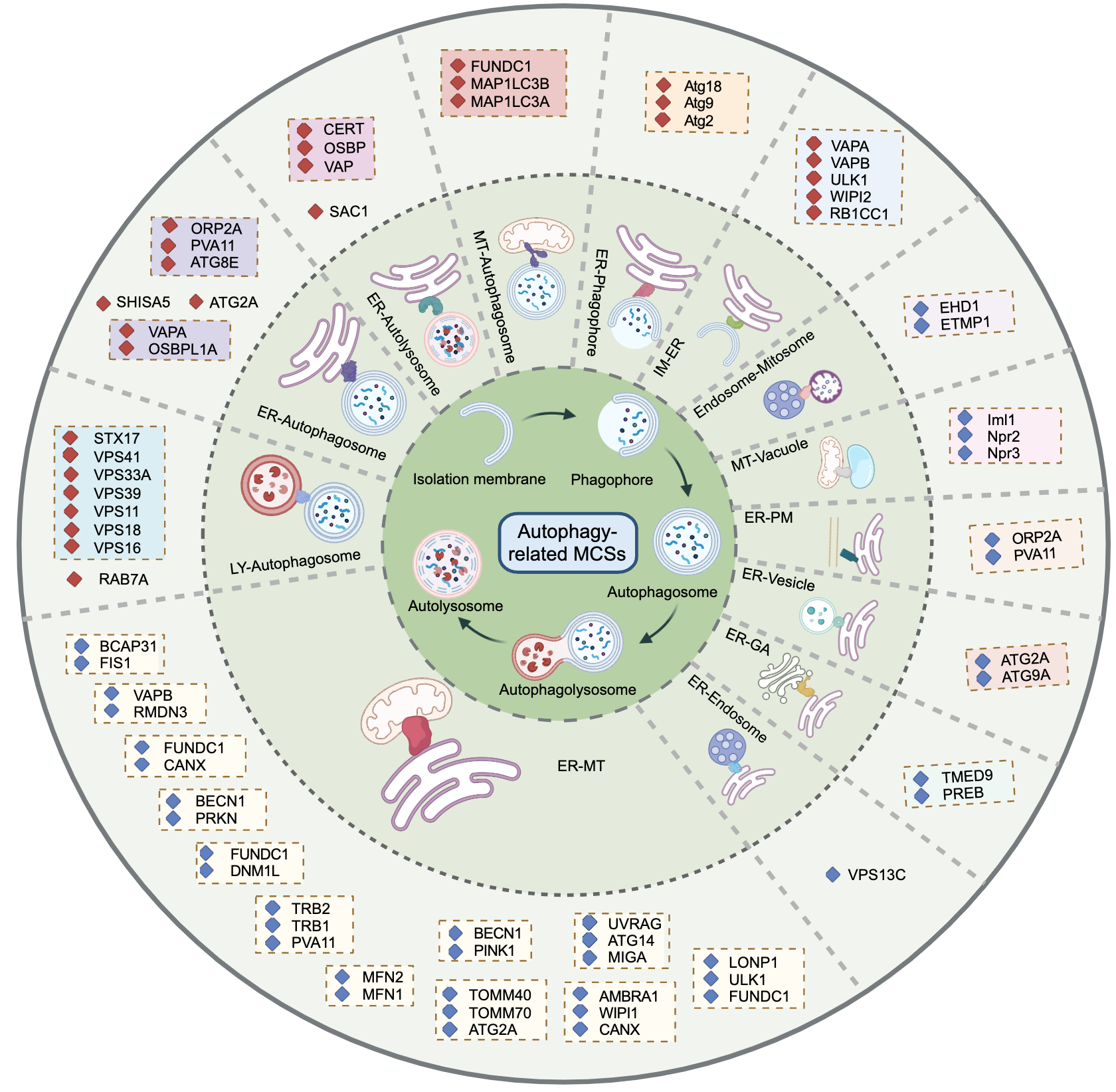
Fig 11-1: Map of autophagy-related MCS protein
Browse by contact site:
As shown in Figure 11-2, user can click on the schematic of the membrane contact sites in the circular diagram.
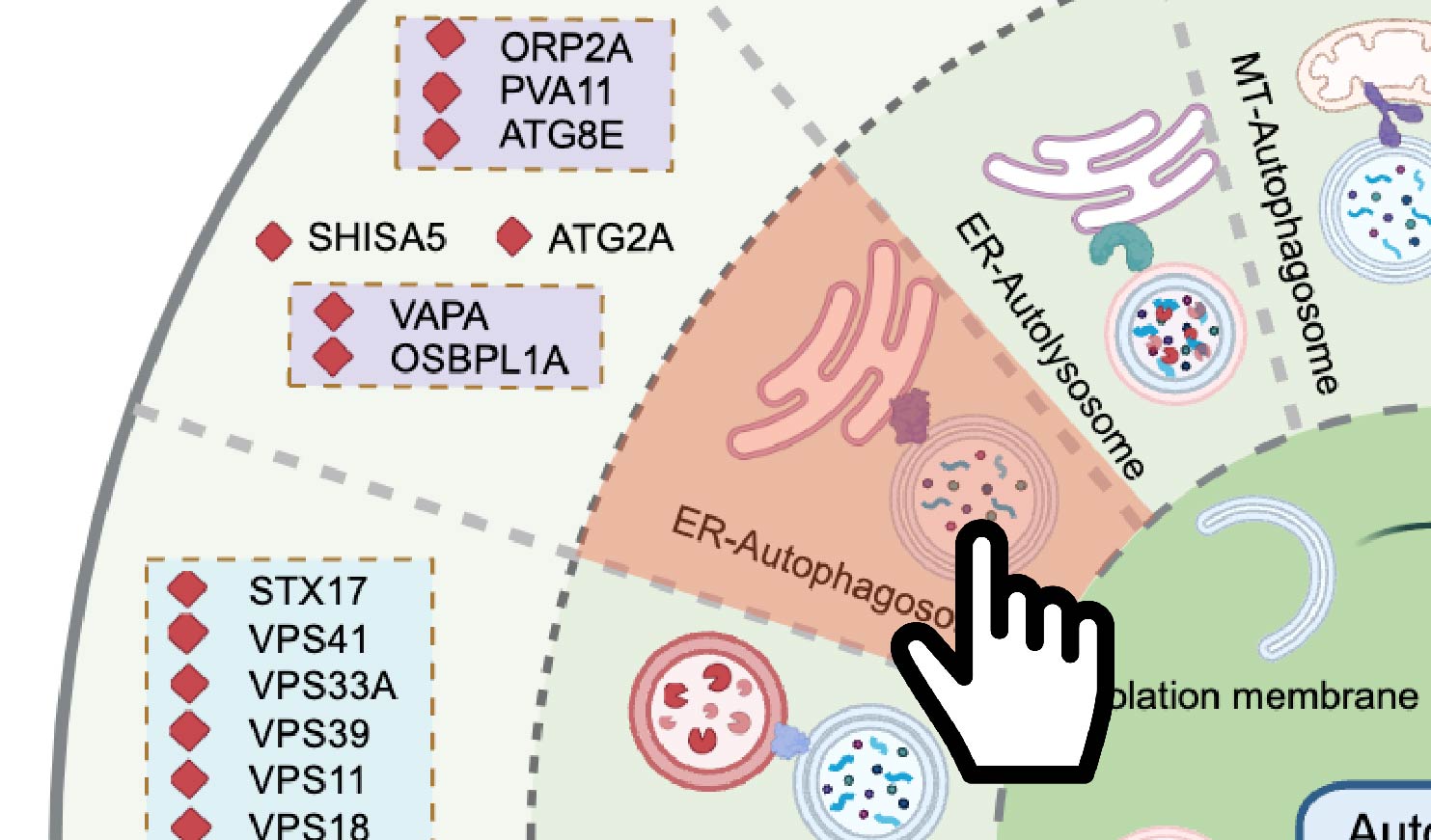
Fig 11-2: Search for membrane contact site
Browse by complex or single protein:
As shown in Figure 11-3, user can click on the schematic of the complex or single protein in the circular diagram.
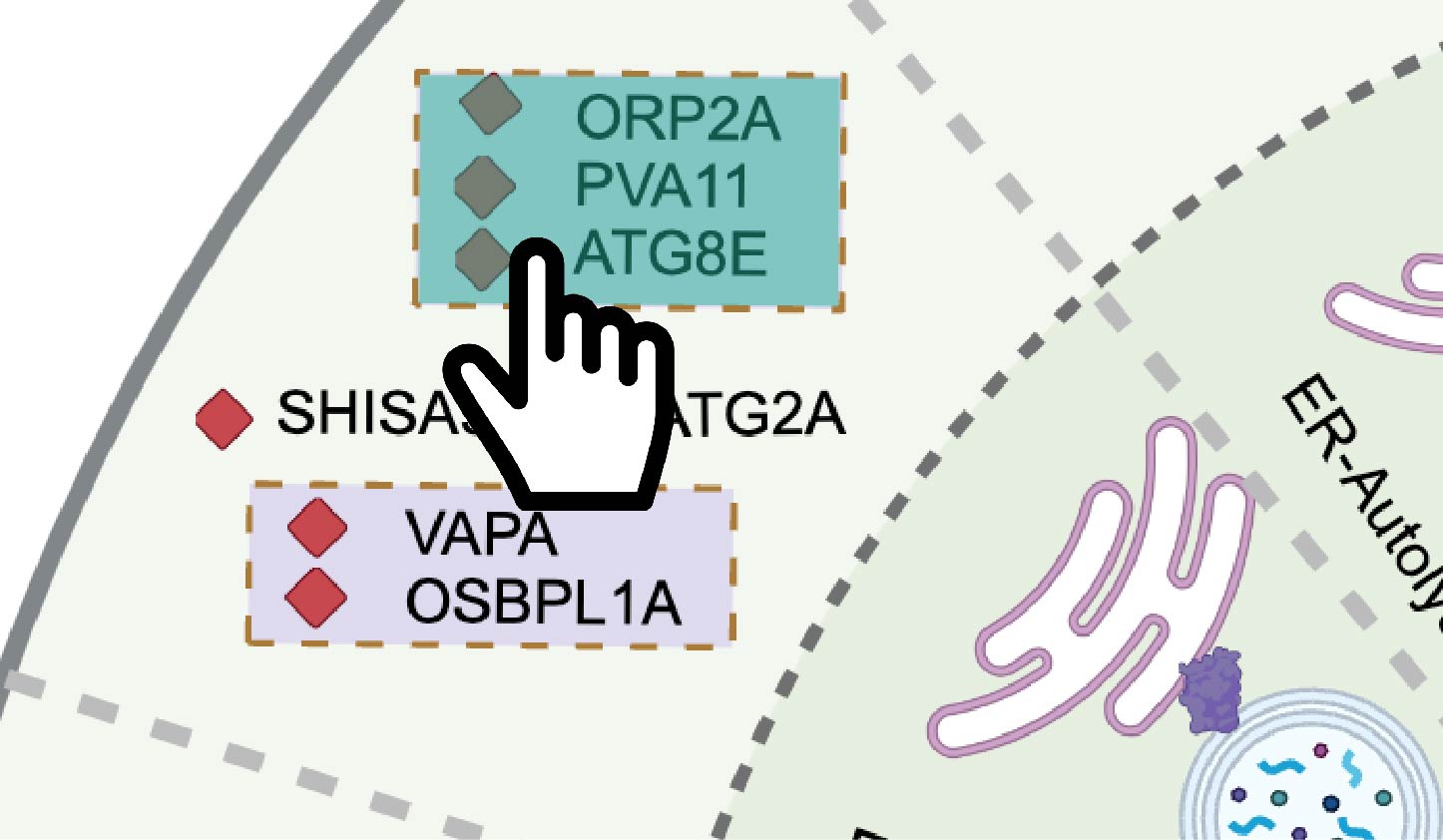
Fig 11-3: Search for complex or single protein