Overview:
The current version of the siRNAEfficacyDB contains more than 3,000 manually collated data records of siRNA drug efficacy, including its sequence and various characteristics. Taken together, siRNAEfficacyDB provides a convenient, user-friendly interface for querying, browsing, and visualizing detailed information about these siRNA proteins. It provides the benchmark data set for computer-aided siRNA prediction and design research, and provides data support for nucleic acid drug research and development.
The homepage is displayed in the following Fig 1-1:
1. Main functions of the database are provided in menu bar form (boxed in red).
2. Introduction and overview for siRNAEfficacyDB.
3. Quick search for users.
4. Declaration of Data Availability.
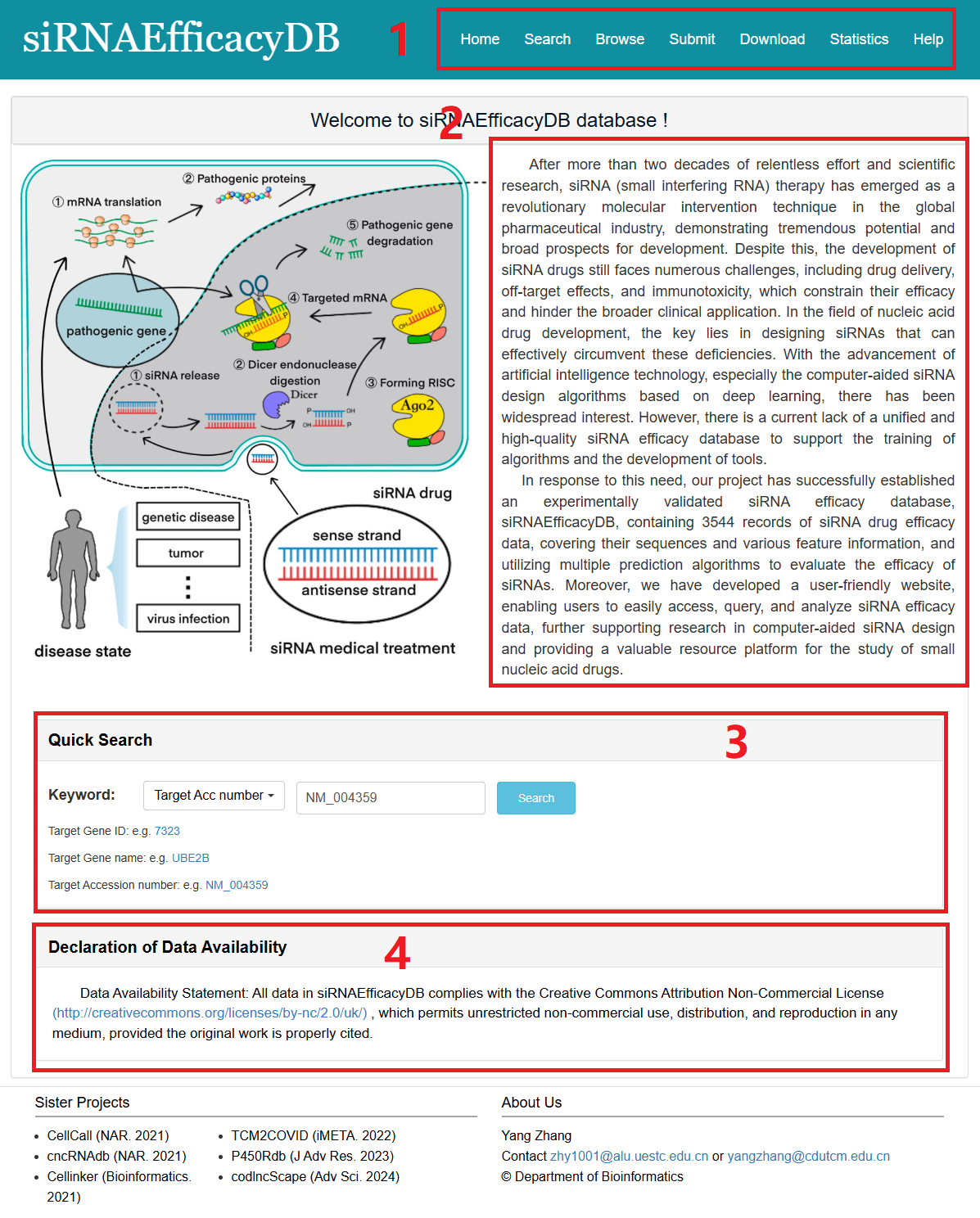
Fig 1-1. Home page
The search page is displayed in Fig 2-1, 2-2 and 2-3:
ID search:
1. Select ID search.
2. Select type of input keyword: three choices are provided (Target Acc number/Target Gene/Target Gene ID).
3. Input a keyword corresponding to selected type.
4. Select the Cell Line to filter the search.
5. Select the Experimental Method to filter the search.
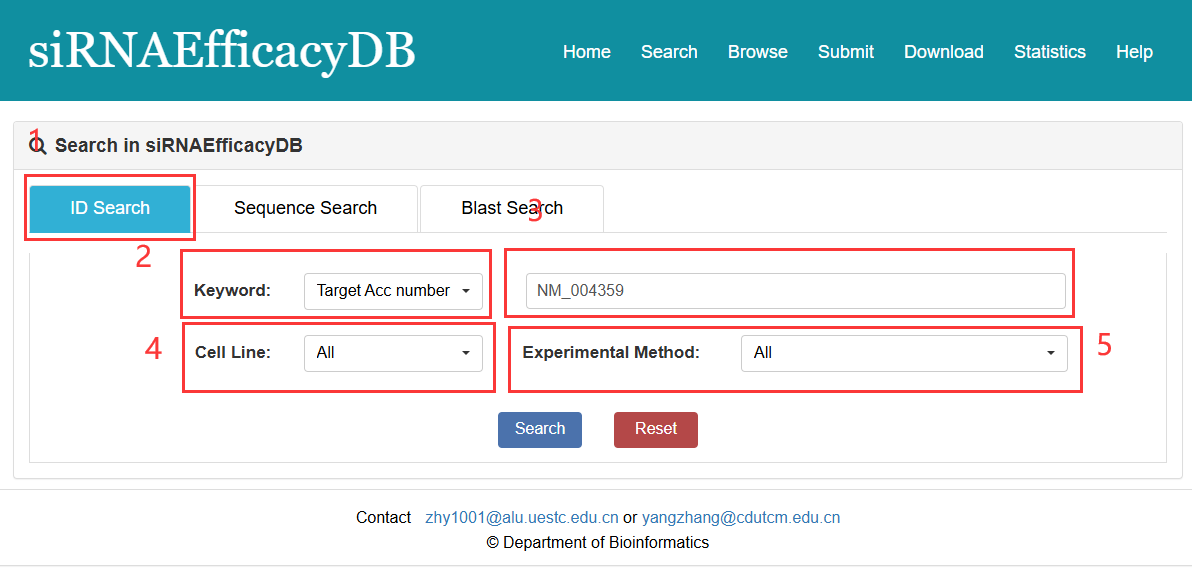
Fig 2-1. ID Search page
Sequence search:
1. Select Sequence search.
2. Select type of input keyword: two choices are provided (Antisense strand/Sense strand).
3. Input list of keywords corresponding to selected type.
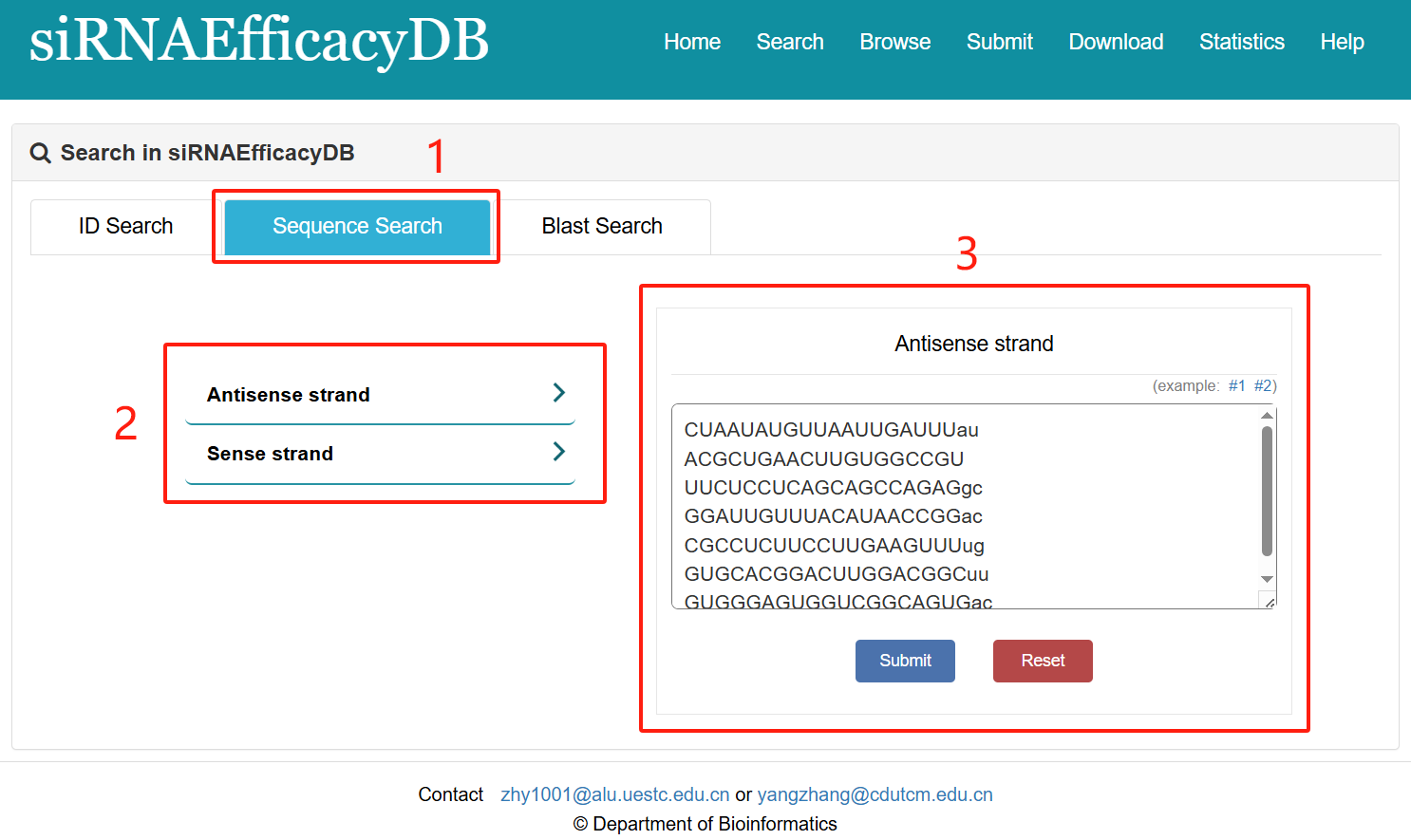
Fig 2-2. Sequence Search page
Blast search:
1. Select Blast search.
2. Enter a sequence to do search.
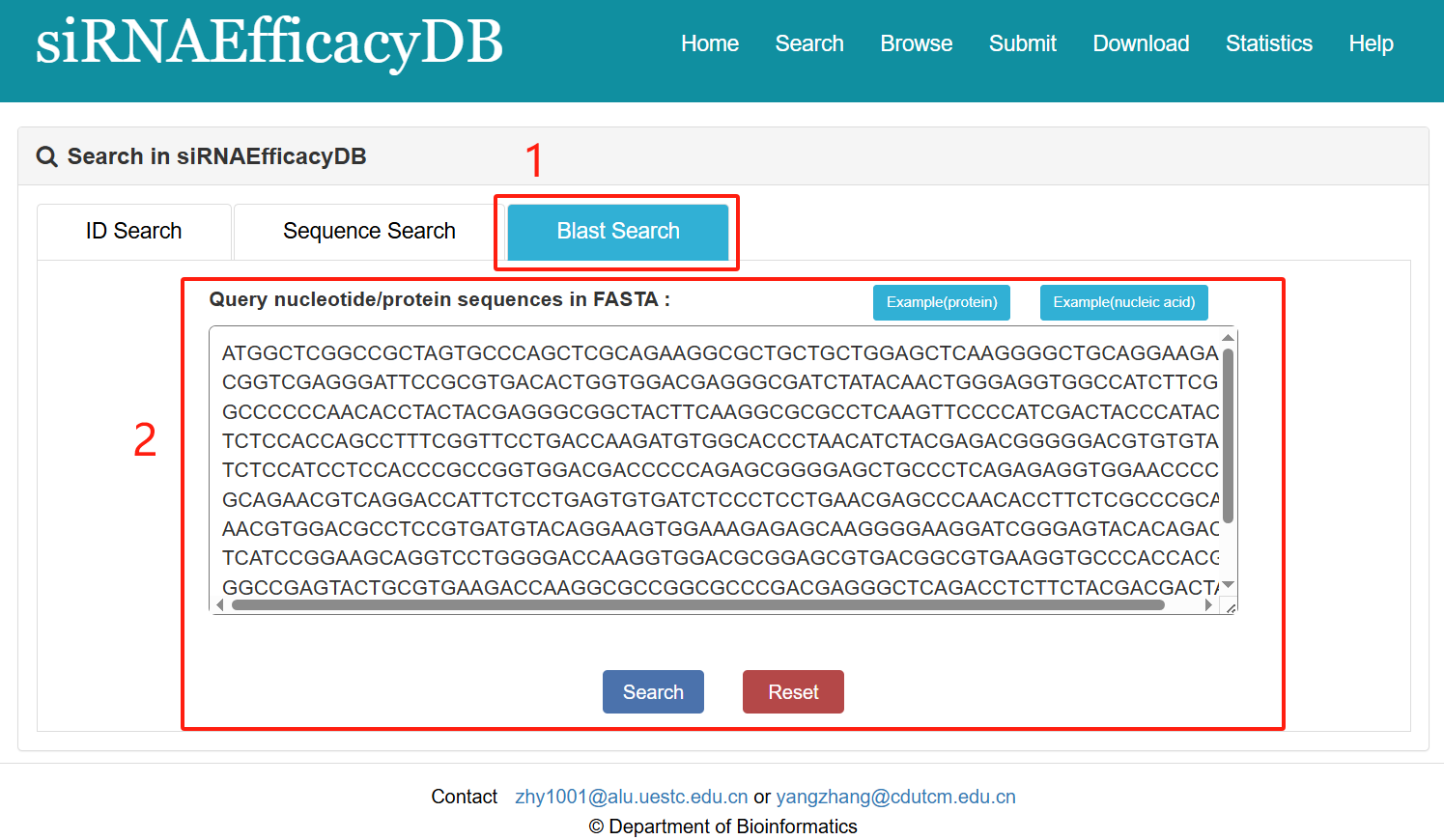
Fig 2-3. Blast Search page
Result page of Exact search and Batch search:
For the result page of Exact search,Sequence features and Batch search, all entries are listed with basic information including Accession number,Gene,Gene ID,Technology,Cell,Concentration,Hours,%Inhibition.
Fig 3-1:
1. Search keyword from the result table.
2. The result table (including Accession number,Gene,Gene ID,Technology,Cell,Concentration,Hours,%Inhibition).
3. Click to link to Detail page.
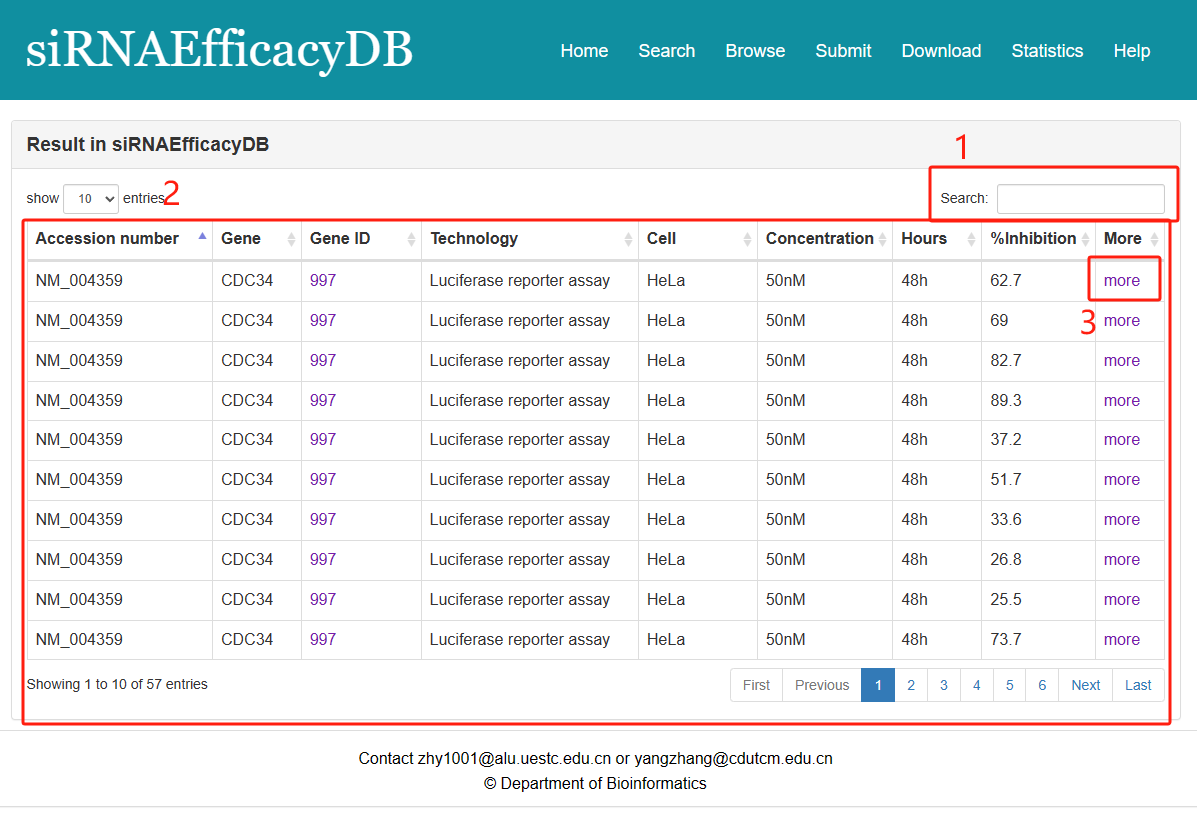
Fig 3-1. Result page of Exact search,Sequence features and Batch search.
In the Detail page, you can get the detail information, including "Basic Information","Characteristics"and "References".
Fig 4-1:
1. Basic Information: including Antisense strand,Sense strand,Sequence Alignment,Target Accession number,Target Gene,Target Gene ID,Technology,Target Cell,Concentration,Hours and %Inhibition.
2. Characteristics: including 5' end, 3' end , Sec.dG, GC stretch,Whole ∆G,%GC.
3.prediction methods: including Hsieh,Amarzguioui,Katoh,Takasaki,Reynolds,Ui-Tei and i-score.
4. The PMID, Title, Journal and published years of the references.

Fig 4-1. Detail page
Users can search for relevant information based on target genes and target cells.
Fig 5-1:
1. Filter by Target Genes (42 type of Genes).
2. Filter by Target Cells (7 type of Cells).
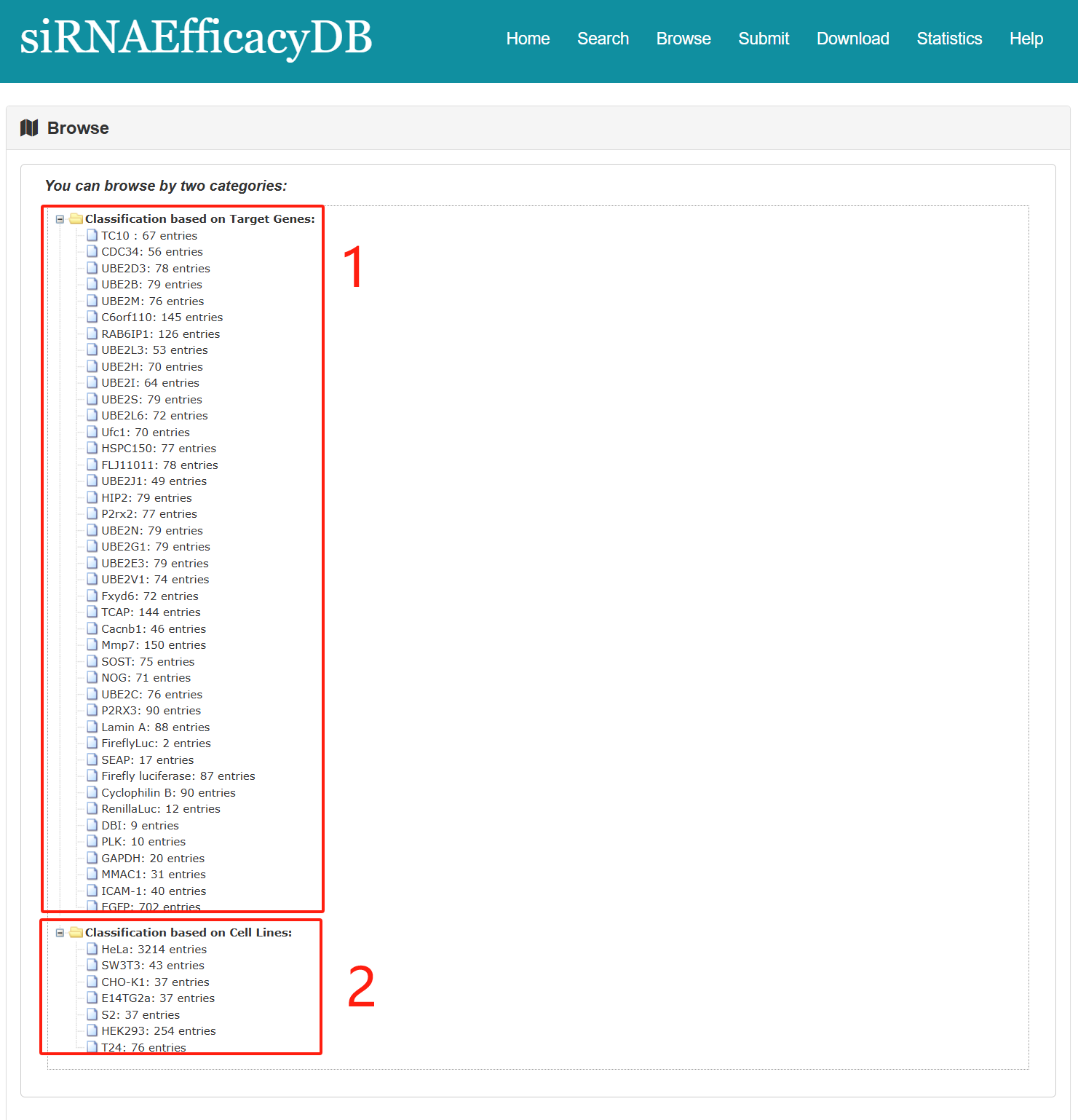
Fig 5-1. Browse page
siRNAEfficacyDB provide the download page for users. You can download all the relevant information in the download page.
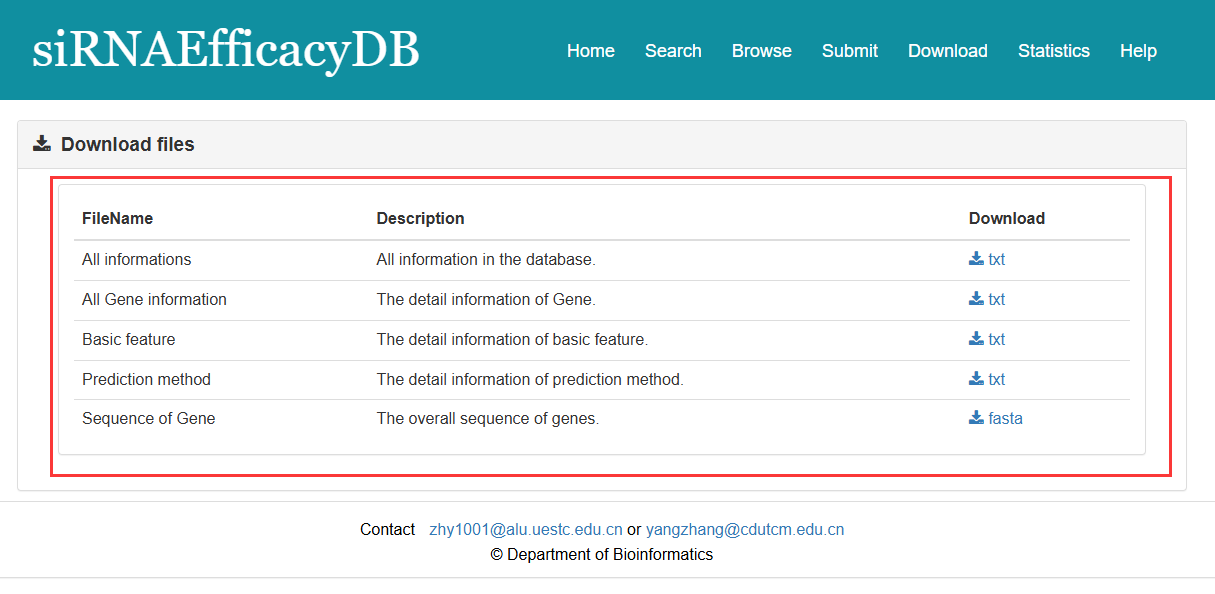
Fig 6-1. Download page